Quantitative Proteome Analysis Reveals Melissa officinalis Extract Targets Mitochondrial Respiration in Colon Cancer Cells
Abstract
:1. Introduction
2. Results
2.1. TMT-Based Quantitative Proteomics Analysis of MO Treatment in HCT116 Cells
2.2. Functional Enrichment Analysis of MO-Regulated Proteins in HCT116 Cells
2.3. MO Decreased Protein Expression Levels of Mitochondrial Complex I, II, IV
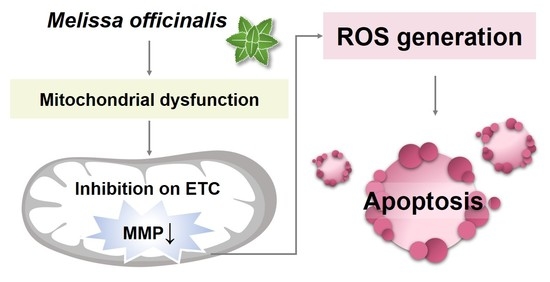
2.4. MO-Induced Apoptosis and Mitochondrial Dysfunction Were Mediated by ROS Production
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. MO Extraction, Chemicals, and Antibodies
4.3. Cell Viability Assay
4.4. Sample Preparation for Mass Spectrometry (MS) Analysis
4.5. TMT Labeling and bRP Fractionation
4.6. LC-MS/MS Analysis
4.7. MS Data Processing
4.8. MS Data Analysis and Functional Enrichment Analysis
4.9. ROS Measurement Assay
4.10. Apoptosis Assay and Mitochondrial Membrane Potential (MMP) Assay
4.11. Western Blot Analysis
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations:
References
- Moradkhani, H.; Sargsyan, E.; Bibak, H.; Naseri, B.; Sadat-Hosseini, M.; Fayazi-Barjin, A.; Meftahizade, H. Melissa officinalis L., a valuable medicine plant: A review. J. Med. Plants Res. 2010, 4, 2753–2759. [Google Scholar]
- Uritu, C.M.; Mihai, C.T.; Stanciu, G.D.; Dodi, G.; Alexa-Stratulat, T.; Luca, A.; Leon-Constantin, M.M.; Stefanescu, R.; Bild, V.; Melnic, S.; et al. Medicinal plants of the family lamiaceae in pain therapy: A review. Pain Res. Manag. 2018, 2018, 7801543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shakeri, A.; Sahebkar, A.; Javadi, B. Melissa officinalis L.—A review of its traditional uses, phytochemistry and pharmacology. J. Ethnopharmacol. 2016, 188, 204–228. [Google Scholar] [CrossRef] [PubMed]
- Sipos, S.; Moaca, E.A.; Pavel, I.Z.; Avram, S.; Cretu, O.M.; Coricovac, D.; Racoviceanu, R.M.; Ghiulai, R.; Pana, R.D.; Soica, C.M.; et al. Melissa officinalis L. Aqueous extract exerts antioxidant and antiangiogenic effects and improves physiological skin parameters. Molecules 2021, 26, 2369. [Google Scholar] [CrossRef]
- Asadi, A.; Shidfar, F.; Safari, M.; Hosseini, A.F.; Fallah Huseini, H.; Heidari, I.; Rajab, A. Efficacy of Melissa officinalis L. (lemon balm) extract on glycemic control and cardiovascular risk factors in individuals with type 2 diabetes: A randomized, double-blind, clinical trial. Phytother. Res. 2019, 33, 651–659. [Google Scholar] [CrossRef]
- Lopez, V.; Martin, S.; Gomez-Serranillos, M.P.; Carretero, M.E.; Jager, A.K.; Calvo, M.I. Neuroprotective and neurological properties of Melissa officinalis. Neurochem. Res. 2009, 34, 1955–1961. [Google Scholar] [CrossRef]
- Naderi Dastjerdi, M.; Darooneh, T.; Nasiri, M.; Moatar, F.; Esmaeili, S.; Ozgoli, G. Investigating the effect of Melissa officinalis on after-pains: A randomized single-blind clinical trial. J. Caring Sci. 2019, 8, 129–138. [Google Scholar] [CrossRef]
- Mazzanti, G.; Battinelli, L.; Pompeo, C.; Serrilli, A.M.; Rossi, R.; Sauzullo, I.; Mengoni, F.; Vullo, V. Inhibitory activity of Melissa officinalis L. Extract on herpes simplex virus type 2 replication. Nat. Prod. Res. 2008, 22, 1433–1440. [Google Scholar] [CrossRef]
- Saraydin, S.U.; Tuncer, E.; Tepe, B.; Karadayi, S.; Ozer, H.; Sen, M.; Karadayi, K.; Inan, D.; Elagoz, S.; Polat, Z.; et al. Antitumoral effects of Melissa officinalis on breast cancer in vitro and in vivo. Asian Pac. J. Cancer Prev. 2012, 13, 2765–2770. [Google Scholar] [CrossRef] [Green Version]
- Weidner, C.; Rousseau, M.; Plauth, A.; Wowro, S.J.; Fischer, C.; Abdel-Aziz, H.; Sauer, S. Melissa officinalis extract induces apoptosis and inhibits proliferation in colon cancer cells through formation of reactive oxygen species. Phytomedicine 2015, 22, 262–270. [Google Scholar] [CrossRef]
- Kuo, T.T.; Chang, H.Y.; Chen, T.Y.; Liu, B.C.; Chen, H.Y.; Hsiung, Y.C.; Hsia, S.M.; Chang, C.J.; Huang, T.C. Melissa officinalis extract induces apoptosis and inhibits migration in human colorectal cancer cells. ACS Omega 2020, 5, 31792–31800. [Google Scholar] [CrossRef]
- Lao, Y.; Wang, X.; Xu, N.; Zhang, H.; Xu, H. Application of proteomics to determine the mechanism of action of traditional chinese medicine remedies. J. Ethnopharmacol. 2014, 155, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Yang, H.L.; Wang, P.; Lu, Y.C.; Yang, Y.J.; Wang, L.; Lee, S.C. Functional proteomic analysis revels that the ethanol extract of annona muricata l. Induces liver cancer cell apoptosis through endoplasmic reticulum stress pathway. J. Ethnopharmacol. 2016, 189, 210–217. [Google Scholar] [CrossRef] [Green Version]
- Ross, P.L.; Huang, Y.N.; Marchese, J.N.; Williamson, B.; Parker, K.; Hattan, S.; Khainovski, N.; Pillai, S.; Dey, S.; Daniels, S.; et al. Multiplexed protein quantitation in saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteom. 2004, 3, 1154–1169. [Google Scholar] [CrossRef] [Green Version]
- Thompson, A.; Schafer, J.; Kuhn, K.; Kienle, S.; Schwarz, J.; Schmidt, G.; Neumann, T.; Johnstone, R.; Mohammed, A.K.; Hamon, C. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by ms/ms. Anal. Chem. 2003, 75, 1895–1904. [Google Scholar] [CrossRef]
- Rauniyar, N.; Yates, J.R., 3rd. Isobaric labeling-based relative quantification in shotgun proteomics. J. Proteome Res. 2014, 13, 5293–5309. [Google Scholar] [CrossRef] [Green Version]
- Batth, T.S.; Francavilla, C.; Olsen, J.V. Off-line high-ph reversed-phase fractionation for in-depth phosphoproteomics. J. Proteome Res. 2014, 13, 6176–6186. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. Maxquant enables high peptide identification rates, individualized p.P.B.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Tyanova, S.; Cox, J. Perseus: A bioinformatics platform for integrative analysis of proteomics data in cancer research. Methods Mol. Biol. 2018, 1711, 133–148. [Google Scholar]
- Tusher, V.G.; Tibshirani, R.; Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 2001, 98, 5116–5121. [Google Scholar] [CrossRef] [Green Version]
- Roxas, B.A.; Li, Q. Significance analysis of microarray for relative quantitation of lc/ms data in proteomics. BMC Bioinform. 2008, 9, 187. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Boyle, E.I.; Weng, S.; Gollub, J.; Jin, H.; Botstein, D.; Cherry, J.M.; Sherlock, G. Go::Termfinder—Open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics 2004, 20, 3710–3715. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.X.; Thompson, K.; Taylor, R.W.; Olahova, M. Mitochondrial oxphos biogenesis: Co-regulation of protein synthesis, import, and assembly pathways. Int. J. Mol. Sci. 2020, 21, 3820. [Google Scholar] [CrossRef]
- Huttemann, M.; Lee, I.; Samavati, L.; Yu, H.; Doan, J.W. Regulation of mitochondrial oxidative phosphorylation through cell signaling. Biochim. Biophys. Acta 2007, 1773, 1701–1720. [Google Scholar] [CrossRef] [Green Version]
- Brenner, C.; Kroemer, G. Apoptosis. Mitochondria—The death signal integrators. Science 2000, 289, 1150–1151. [Google Scholar] [CrossRef]
- Sivandzade, F.; Bhalerao, A.; Cucullo, L. Analysis of the mitochondrial membrane potential using the cationic jc-1 dye as a sensitive fluorescent probe. Bio Protoc. 2019, 9, e3128. [Google Scholar] [CrossRef]
- Suski, J.M.; Lebiedzinska, M.; Bonora, M.; Pinton, P.; Duszynski, J.; Wieckowski, M.R. Relation between mitochondrial membrane potential and ros formation. Methods Mol. Biol. 2012, 810, 183–205. [Google Scholar]
- Zou, Z.; Chang, H.; Li, H.; Wang, S. Induction of reactive oxygen species: An emerging approach for cancer therapy. Apoptosis 2017, 22, 1321–1335. [Google Scholar] [CrossRef]
- Lebiedzinska, M.; Karkucinska-Wieckowska, A.; Giorgi, C.; Karczmarewicz, E.; Pronicka, E.; Pinton, P.; Duszynski, J.; Pronicki, M.; Wieckowski, M.R. Oxidative stress-dependent p66shc phosphorylation in skin fibroblasts of children with mitochondrial disorders. Biochim. Biophys. Acta 2010, 1797, 952–960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panieri, E.; Santoro, M.M. Ros homeostasis and metabolism: A dangerous liason in cancer cells. Cell Death Dis. 2016, 7, e2253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magalhaes, D.B.; Castro, I.; Lopes-Rodrigues, V.; Pereira, J.M.; Barros, L.; Ferreira, I.; Xavier, C.P.R.; Vasconcelos, M.H. Melissa officinalis L. Ethanolic extract inhibits the growth of a lung cancer cell line by interfering with the cell cycle and inducing apoptosis. Food Funct. 2018, 9, 3134–3142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Encalada, M.A.; Hoyos, K.M.; Rehecho, S.; Berasategi, I.; de Ciriano, M.G.; Ansorena, D.; Astiasaran, I.; Navarro-Blasco, I.; Cavero, R.Y.; Calvo, M.I. Anti-proliferative effect of Melissa officinalis on human colon cancer cell line. Plant Foods Hum. Nutr. 2011, 66, 328–334. [Google Scholar] [CrossRef]
- Ma, Z.; Yang, J.; Yang, Y.; Wang, X.; Chen, G.; Shi, A.; Lu, Y.; Jia, S.; Kang, X.; Lu, L. Rosmarinic acid exerts an anticancer effect on osteosarcoma cells by inhibiting dj-1 via regulation of the pten-pi3k-akt signaling pathway. Phytomedicine 2020, 68, 153186. [Google Scholar] [CrossRef]
- Pu, Y.S.; Zhang, T.; Wang, J.H.; Mao, Z.J.; Duan, B.J.; Long, Y.B.; Xue, F.; Liu, D.; Liu, S.D.; Gao, Z.Z. Luteolin exerts an anticancer effect on gastric cancer cells through multiple signaling pathways and regulating mirnas. J. Cancer 2018, 9, 3669–3675. [Google Scholar] [CrossRef] [Green Version]
- Kuhlbrandt, W. Structure and function of mitochondrial membrane protein complexes. BMC Biol. 2015, 13, 89. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.T.; Huang, H.C.; Lin, J.K. Rotenone induces apoptosis in mcf-7 human breast cancer cell-mediated ros through jnk and p38 signaling. Mol. Carcinog. 2010, 49, 141–151. [Google Scholar] [CrossRef]
- Kim, S.J.; Kim, H.S.; Seo, Y.R. Understanding of ros-inducing strategy in anticancer therapy. Oxid. Med. Cell Longev. 2019, 2019, 5381692. [Google Scholar] [CrossRef]
- Lee, W.L.; Huang, J.Y.; Shyur, L.F. Phytoagents for cancer management: Regulation of nucleic acid oxidation, ros, and related mechanisms. Oxid. Med. Cell Longev. 2013, 2013, 925804. [Google Scholar] [CrossRef]
- Simon, H.U.; Haj-Yehia, A.; Levi-Schaffer, F. Role of reactive oxygen species (ros) in apoptosis induction. Apoptosis 2000, 5, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Diez, C.; Miguel, V.; Mennerich, D.; Kietzmann, T.; Sanchez-Perez, P.; Cadenas, S.; Lamas, S. Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol. 2015, 6, 183–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorrini, C.; Harris, I.S.; Mak, T.W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 2013, 12, 931–947. [Google Scholar] [CrossRef] [PubMed]
- Borrero, L.J.H.; El-Deiry, W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta BBA—Rev. Cancer 2021, 1876, 188556. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Hu, W.; Feng, Z. Tumor suppressor p53 and metabolism. J. Mol. Cell. Biol. 2019, 11, 284–292. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Qian, Y.; Zhang, J.; Yan, W.; Jung, Y.-S.; Chen, M.; Huang, E.; Lloyd, K.; Duan, Y.; Wang, J. Ferredoxin reductase is critical for p53-dependent tumor suppression via iron regulatory protein 2. Genes Dev. 2017, 31, 1243–1256. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Chen, X. The ferredoxin reductase gene is regulated by the p53 family and sensitizes cells to oxidative stress-induced apoptosis. Oncogene 2002, 21, 7195–7204. [Google Scholar] [CrossRef] [Green Version]
- Phan, L.; Chou, P.-C.; Velazquez-Torres, G.; Samudio, I.; Parreno, K.; Huang, Y.; Tseng, C.; Vu, T.; Gully, C.; Su, C.-H. The cell cycle regulator 14-3-3σ opposes and reverses cancer metabolic reprogramming. Nat. Commun. 2015, 6, 7530. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Comish, P.B.; Tang, D.; Kang, R. Characteristics and biomarkers of ferroptosis. Front. Cell Dev. Biol. 2021, 9, 637162. [Google Scholar] [CrossRef]
- Chiang, S.-K.; Chen, S.-E.; Chang, L.-C. A dual role of heme oxygenase-1 in cancer cells. Int. J. Mol. Sci. 2018, 20, 39. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.J.; Li, D.; Ou, Y.; Jiang, L.; Chen, Y.; Zhao, Y.; Gu, W. Acetylation is crucial for p53-mediated ferroptosis and tumor suppression. Cell Rep. 2016, 17, 366–373. [Google Scholar] [CrossRef] [Green Version]
- Kan, X.; Yin, Y.; Song, C.; Tan, L.; Qiu, X.; Liao, Y.; Liu, W.; Meng, S.; Sun, Y.; Ding, C. Newcastle-disease-virus-induced ferroptosis through nutrient deprivation and ferritinophagy in tumor cells. iScience 2021, 24, 102837. [Google Scholar] [CrossRef]
- Masuda, T.; Tomita, M.; Ishihama, Y. Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J. Proteome Res. 2008, 7, 731–740. [Google Scholar] [CrossRef]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using stagetips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the maxquant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Chawade, A.; Alexandersson, E.; Levander, F. Normalyzer: A tool for rapid evaluation of normalization methods for omics data sets. J. Proteome Res. 2014, 13, 3114–3120. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative html5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [Green Version]
- Moriya, Y.; Kawano, S.; Okuda, S.; Watanabe, Y.; Matsumoto, M.; Takami, T.; Kobayashi, D.; Yamanouchi, Y.; Araki, N.; Yoshizawa, A.C. The jpost environment: An integrated proteomics data repository and database. Nucleic Acids Res. 2019, 47, D1218–D1224. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuo, T.-T.; Lin, L.-C.; Chang, H.-Y.; Chiang, P.-J.; Wu, H.-Y.; Chen, T.-Y.; Hsia, S.-M.; Huang, T.-C. Quantitative Proteome Analysis Reveals Melissa officinalis Extract Targets Mitochondrial Respiration in Colon Cancer Cells. Molecules 2022, 27, 4533. https://doi.org/10.3390/molecules27144533
Kuo T-T, Lin L-C, Chang H-Y, Chiang P-J, Wu H-Y, Chen T-Y, Hsia S-M, Huang T-C. Quantitative Proteome Analysis Reveals Melissa officinalis Extract Targets Mitochondrial Respiration in Colon Cancer Cells. Molecules. 2022; 27(14):4533. https://doi.org/10.3390/molecules27144533
Chicago/Turabian StyleKuo, Tzu-Ting, Li-Chun Lin, Hsin-Yi Chang, Pei-Jung Chiang, Hsin-Yi Wu, Tai-Yuan Chen, Shih-Min Hsia, and Tsui-Chin Huang. 2022. "Quantitative Proteome Analysis Reveals Melissa officinalis Extract Targets Mitochondrial Respiration in Colon Cancer Cells" Molecules 27, no. 14: 4533. https://doi.org/10.3390/molecules27144533
APA StyleKuo, T. -T., Lin, L. -C., Chang, H. -Y., Chiang, P. -J., Wu, H. -Y., Chen, T. -Y., Hsia, S. -M., & Huang, T. -C. (2022). Quantitative Proteome Analysis Reveals Melissa officinalis Extract Targets Mitochondrial Respiration in Colon Cancer Cells. Molecules, 27(14), 4533. https://doi.org/10.3390/molecules27144533