Mass Spectrometry-Based Peptide Profiling of Haemolymph from Pterostichus melas Exposed to Pendimethalin Herbicide
Abstract
:1. Introduction
2. Results and Discussion
2.1. MALDI MS Metabolites Identification
2.2. Bioinformatic Analysis
2.3. Chemical Component Profile of Control and PND-Treated Groups
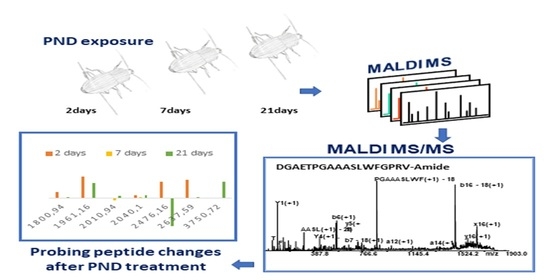
2.4. Probing Peptide Changes after PND Treatment
3. Materials and Methods
3.1. Sample Collection and Treatment
3.2. Haemolymph Collection
3.3. Sample Preparation
3.4. MALDI MS Analysis
3.5. Database Proteomics and Targeting Predictions
3.6. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Vighi, M.; Matthies, M.; Solomon, K.R. Critical Assessment of Pendimethalin in Terms of Persistence, Bioaccumulation, Toxicity, and Potential for Long-Range Transport. J. Toxicol. Environ. Health Part B 2017, 20, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strandberg, M.; Scott-Fordsmand, J.J. Effects of Pendimethalin at Lower Trophic Levels—A Review. Ecotoxicol. Environ. Saf. 2004, 57, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Roca, E.; D’Errico, E.; Izzo, A.; Strumia, S.; Esposito, A.; Fiorentino, A. In Vitro Saprotrophic Basidiomycetes Tolerance to Pendimethalin. Int. Biodeterior. Biodegrad. 2009, 63, 182–186. [Google Scholar] [CrossRef]
- Kočárek, M.; Artikov, H.; Voříšek, K.; Borůvka, L. Pendimethalin Degradation in Soil and Its Interaction with Soil Microorganisms. Soil Water Res. 2016, 11, 213–219. [Google Scholar] [CrossRef] [Green Version]
- Belden, J.B.; Phillips, T.A.; Clark, B.W.; Coats, J.R. Toxicity of Pendimethalin to Nontarget Soil Organisms. Bull. Environ. Contam. Toxicol. 2005, 74, 769–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliver, J.B.; Reding, M.E.; Moyseenko, J.J.; Klein, M.G.; Mannion, C.M.; Bishop, B. Survival of Adult Tiphia vernalis (Hymenoptera: Tiphiidae) After Insecticide, Fungicide, and Herbicide Exposure in Laboratory Bioassays. J. Econ. Entomol. 2006, 99, 288–294. [Google Scholar] [CrossRef]
- Giglio, A.; Cavaliere, F.; Giulianini, P.G.; Kurtz, J.; Vommaro, M.L.; Brandmayr, P. Continuous Agrochemical Treatments in Agroecosystems Can Modify the Effects of Pendimethalin-Based Herbicide Exposure on Immunocompetence of a Beneficial Ground Beetle. Diversity 2019, 11, 241. [Google Scholar] [CrossRef] [Green Version]
- De Heij, S.E.; Willenborg, C.J. Connected Carabids: Network Interactions and Their Impact on Biocontrol by Carabid Beetles. BioScience 2020, 70, 490–500. [Google Scholar] [CrossRef]
- Giglio, A.; Brandmayr, P.; Talarico, F.; Giulianini, P.G. Effects of alternative and specialised diet on development and survival of larvae and pupae in Carabus (Chaetocarabus) lefebvrei (Coleoptera: Carabidae). Entomol. Gen. 2012, 33, 263–271. [Google Scholar] [CrossRef]
- Foffová, H.; Ćavar Zeljković, S.; Honěk, A.; Martinková, Z.; Tarkowski, P.; Saska, P. Which seed properties determine the preferences of carabid beetle seed predators? Insects 2020, 11, 757. [Google Scholar] [CrossRef]
- Giglio, A.; Cavaliere, F.; Giulianini, P.G.; Mazzei, A.; Talarico, F.; Vommaro, M.L.; Brandmayr, P. Impact of Agrochemicals on Non-Target Species: Calathus Fuscipes Goeze 1777 (Coleoptera: Carabidae) as Model. Ecotoxicol. Environ. Saf. 2017, 142, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Holland, J.M.; Luff, M.L. The Effects of Agricultural Practices on Carabidae in Temperate Agroecosystems. Integr. Pest Manag. Rev. 2000, 5, 109–129. [Google Scholar] [CrossRef]
- Cavaliere, F.; Brandmayr, P.; Giglio, A. DNA Damage in Haemocytes of Harpalus (Pseudophonus) Rufipes (De Geer, 1774) (Coleoptera, Carabidae) as an Indicator of Sublethal Effects of Exposure to Herbicides. Ecol. Indic. 2019, 98, 88–91. [Google Scholar] [CrossRef]
- Prosser, R.S.; Anderson, J.C.; Hanson, M.L.; Solomon, K.R.; Sibley, P.K. Indirect Effects of Herbicides on Biota in Terrestrial Edge-of-Field Habitats: A Critical Review of the Literature. Agric. Ecosyst. Environ. 2016, 232, 59–72. [Google Scholar] [CrossRef]
- Alonso, A.; Marsal, S.; Julià, A. Analytical Methods in Untargeted Metabolomics: State of the Art in 2015. Front. Bioeng. Biotechnol. 2015, 3, 23. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Li, Q.; Xu, Z.; Dou, J. Mass Spectrometry-Based Metabolomics in Health and Medical Science: A Systematic Review. RSC Adv. 2020, 10, 3092–3104. [Google Scholar] [CrossRef] [Green Version]
- Chapman, R.F.; Simpson, S.J.; Douglas, A.E. The Insects: Structure and Function, 5th ed.; Cambridge University Press: New York, NY, USA, 2013. [Google Scholar]
- Rosales, C. Cellular and Molecular Mechanisms of Insect Immunity. In Insect Physiology and Ecology; Shields, V.D.C., Ed.; InTech: Rijeka, Croatia, 2017. [Google Scholar] [CrossRef] [Green Version]
- Aliferis, K.A.; Copley, T.; Jabaji, S. Gas Chromatography–Mass Spectrometry Metabolite Profiling of Worker Honey Bee (Apis mellifera L.) Hemolymph for the Study of Nosema Ceranae Infection. J. Insect Physiol. 2012, 58, 1349–1359. [Google Scholar] [CrossRef]
- Aiello, D.; Giambona, A.; Leto, F.; Passarello, C.; Damiani, G.; Maggio, A.; Siciliano, C.; Napoli, A. Human Coelomic Fluid Investigation: A MS-Based Analytical Approach to Prenatal Screening. Sci. Rep. 2018, 8, 10973. [Google Scholar] [CrossRef] [Green Version]
- Imbrogno, S.; Aiello, D.; Filice, M.; Leo, S.; Mazza, R.; Cerra, M.C.; Napoli, A. MS-Based Proteomic Analysis of Cardiac Response to Hypoxia in the Goldfish (Carassius auratus). Sci. Rep. 2019, 9, 18953. [Google Scholar] [CrossRef] [Green Version]
- Chavy, A.; Nabet, C.; Normand, A.C.; Kocher, A.; Ginouves, M.; Prévot, G.; Vasconcelos dos Santos, T.; Demar, M.; Piarroux, R.; de Thoisy, B. Identification of French Guiana Sand Flies Using MALDI-TOF Mass Spectrometry with a New Mass Spectra Library. PLoS Negl. Trop. Dis. 2019, 13, e0007031. [Google Scholar] [CrossRef]
- Vega-Rúa, A.; Pagès, N.; Fontaine, A.; Nuccio, C.; Hery, L.; Goindin, D.; Gustave, J.; Almeras, L. Improvement of Mosquito Identification by MALDI-TOF MS Biotyping Using Protein Signatures from Two Body Parts. Parasites Vectors 2018, 11, 574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, Y.; Zou, Y.; Wang, F.; Gong, J.; Zhong, X.; Xia, Q.; Zhao, P. Comparative Analysis of Proteome Maps of Silkworm Hemolymph during Different Developmental Stages. Proteome Sci. 2010, 8, 45. [Google Scholar] [CrossRef] [Green Version]
- Arafah, K.; Voisin, S.N.; Masson, V.; Alaux, C.; Le Conte, Y.; Bocquet, M.; Bulet, P. MALDI–MS Profiling to Address Honey Bee Health Status under Bacterial Challenge through Computational Modeling. Proteomics 2019, 19, 1900268. [Google Scholar] [CrossRef]
- Dani, F.R.; Francese, S.; Mastrobuoni, G.; Felicioli, A.; Caputo, B.; Simard, F.; Pieraccini, G.; Moneti, G.; Coluzzi, M.; della Torre, A.; et al. Exploring Proteins in Anopheles Gambiae Male and Female Antennae through MALDI Mass Spectrometry Profiling. PLoS ONE 2008, 3, e2822. [Google Scholar] [CrossRef] [PubMed]
- Verhaert, P.D.; Conaway, M.C.P.; Pekar, T.M.; Miller, K. Neuropeptide Imaging on an LTQ with VMALDI Source: The Complete ‘All-in-One’ Peptidome Analysis. Int. J. Mass Spectrom. 2007, 260, 177–184. [Google Scholar] [CrossRef]
- Wegener, C.; Reinl, T.; Jänsch, L.; Predel, R. Direct Mass Spectrometric Peptide Profiling and Fragmentation of Larval Peptide Hormone Release Sites in Drosophila Melanogaster Reveals Tagma-Specific Peptide Expression and Differential Processing: Peptidomics of Hormone Release Sites in Drosophila. J. Neurochem. 2006, 96, 1362–1374. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, T.; Mandal, S.M.; Dutta, S.; Ghosh, A.K. Identification of a Novel Proline-rich Antimicrobial Protein from the Hemolymph of Antheraea Mylitta. Arch. Insect Biochem. Physiol. 2021, 106, e21771. [Google Scholar] [CrossRef]
- Catae, A.F.; da Silva Menegasso, A.R.; Pratavieira, M.; Palma, M.S.; Malaspina, O.; Roat, T.C. MALDI-Imaging Analyses of Honeybee Brains Exposed to a Neonicotinoid Insecticide: MALDI-Imaging of Honeybee Brains. Pest Manag. Sci. 2019, 75, 607–615. [Google Scholar] [CrossRef]
- Roat, T.C.; dos Santos-Pinto, J.R.A.; dos Santos, L.D.; Santos, K.S.; Malaspina, O.; Palma, M.S. Modification of the Brain Proteome of Africanized Honeybees (Apis mellifera) Exposed to a Sub-lethal Doses of the Insecticide Fipronil. Ecotoxicology 2014, 23, 1659–1670. [Google Scholar] [CrossRef]
- Thornton, B.J.; Elthon, T.E.; Cerny, R.L.; Siegfried, B.D. Proteomic Analysis of Atrazine Exposure in Drosophila melanogaster (Diptera: Drosophilidae). Chemosphere 2010, 81, 235–241. [Google Scholar] [CrossRef]
- Aiello, D.; Lucà, F.; Siciliano, C.; Frati, P.; Fineschi, V.; Rongo, R.; Napoli, A. Analytical Strategy for MS-Based Thanatochemistry to Estimate Postmortem Interval. J. Proteome Res. 2021, 20, 2607–2617. [Google Scholar] [CrossRef] [PubMed]
- Sunderland, K.D. Invertebrate Pest Control by Carabids. In The Agroecology of Carabid Beetles; Holland, J.M., Ed.; Intercept Limited: Andover, UK, 2002; pp. 165–214. [Google Scholar]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING Database in 2011: Functional Interaction Networks of Proteins, Globally Integrated and Scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef] [PubMed]
- Shiomi, K. Pyrokinin. In Handbook of Hormones; Elsevier: Amsterdam, The Netherlands, 2021; pp. 729–730. [Google Scholar] [CrossRef]
- Lajevardi, A.; Paluzzi, J.-P.V. Receptor Characterization and Functional Activity of Pyrokinins on the Hindgut in the Adult Mosquito, Aedes Aegypti. Front. Physiol. 2020, 11, 490. [Google Scholar] [CrossRef]
- Holman, G.M.; Cook, B.J.; Nachman, R.J. Primary Structure and Synthesis of a Blocked Myotropic Neuropeptide Isolated from the Cockroach, Leucophaea Maderae. Comp. Biochem. Physiol. Part C Comp. Pharmacol. 1986, 85, 219–224. [Google Scholar] [CrossRef]
- Raina, A.K.; Menn, J.J. Pheromone Biosynthesis Activating Neuropeptide: From Discovery to Current Status. Arch. Insect Biochem. Physiol. 1993, 22, 141–151. [Google Scholar] [CrossRef]
- Zhang, Q.; Nachman, R.J.; Kaczmarek, K.; Zabrocki, J.; Denlinger, D.L. Disruption of Insect Diapause Using Agonists and an Antagonist of Diapause Hormone. Proc. Natl. Acad. Sci. USA 2011, 108, 16922–16926. [Google Scholar] [CrossRef] [Green Version]
- Sedra, L.; Lange, A.B. The Female Reproductive System of the Kissing Bug, Rhodnius prolixus: Arrangements of Muscles, Distribution and Myoactivity of Two Endogenous FMRFamide-like Peptides. Peptides 2014, 53, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Suggs, J.M.; Jones, T.H.; Murphree, C.S.; Hillyer, J.F. CCAP and FMRFamide-like Peptides Accelerate the Contraction Rate of the Antennal Accessory Pulsatile Organs (Auxiliary Hearts) of Mosquitoes. J. Exp. Biol. 2016, 219, 2388–2395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenz, O.; Xiong, J.; Nelson, M.D.; Raizen, D.M.; Williams, J.A. FMRFamide Signaling Promotes Stress-Induced Sleep in Drosophila. Brain Behav. Immun. 2015, 47, 141–148. [Google Scholar] [CrossRef] [Green Version]
- Sedra, L.; Lange, A.B. Cloning and Expression of Long Neuropeptide F and the Role of FMRFamide-like Peptides in Regulating Egg Production in the Chagas Vector, Rhodnius prolixus. Peptides 2016, 82, 1–11. [Google Scholar] [CrossRef]
- Hillyer, J.F. Insect Heart Rhythmicity Is Modulated by Evolutionarily Conserved Neuropeptides and Neurotransmitters. Curr. Opin. Insect Sci. 2018, 29, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Marciniak, P.; Witek, W.; Szymczak, M.; Pacholska-Bogalska, J.; Chowański, S.; Kuczer, M.; Rosiński, G. FMRFamide-Related Peptides Signaling Is Involved in the Regulation of Muscle Contractions in Two Tenebrionid Beetles. Front. Physiol. 2020, 11, 456. [Google Scholar] [CrossRef] [PubMed]
- Abou El Asrar, R.; Cools, D.; Vanden Broeck, J. Role of Peptide Hormones in Insect Gut Physiology. Curr. Opin. Insect Sci. 2020, 41, 71–78. [Google Scholar] [CrossRef]
- Rudwall, A.J.; Sliwowska, J.; Nassel, D.R. Allatotropin-like Neuropeptide in the Cockroach Abdominal Nervous System: Myotropic Actions, Sexually Dimorphic Distribution and Colocalization with Serotonin. J. Comp. Neurol. 2000, 428, 159–173. [Google Scholar] [CrossRef]
- Lubawy, J.; Marciniak, P.; Kuczer, M.; Rosiński, G. Myotropic Activity of Allatostatins in Tenebrionid Beetles. Neuropeptides 2018, 70, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Lubawy, J.; Urbański, A.; Colinet, H.; Pflüger, H.-J.; Marciniak, P. Role of the Insect Neuroendocrine System in the Response to Cold Stress. Front. Physiol. 2020, 11, 376. [Google Scholar] [CrossRef]
- Li, B.; Predel, R.; Neupert, S.; Hauser, F.; Tanaka, Y.; Cazzamali, G.; Williamson, M.; Arakane, Y.; Verleyen, P.; Schoofs, L.; et al. Genomics, Transcriptomics, and Peptidomics of Neuropeptides and Protein Hormones in the Red Flour Beetle Tribolium castaneum. Genome Res. 2008, 18, 113–122. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Patočka, J.; Kuča, K. Insect Antimicrobial Peptides, a Mini Review. Toxins 2018, 10, 461. [Google Scholar] [CrossRef]
- Stöcklin, R.; Favreau, P.; Thai, R.; Pflugfelder, J.; Bulet, P.; Mebs, D. Structural Identification by Mass Spectrometry of a Novel Antimicrobial Peptide from the Venom of the Solitary Bee Osmia rufa (Hymenoptera: Megachilidae). Toxicon 2010, 55, 20–27. [Google Scholar] [CrossRef]
- Grosch, J.; Hilger, C.; Bilò, M.B.; Kler, S.; Schiener, M.; Dittmar, G.; Bernardin, F.; Lesur, A.; Ollert, M.; Schmidt-Weber, C.B.; et al. Shedding Light on the Venom Proteomes of the Allergy-Relevant Hymenoptera Polistes dominula (European Paper Wasp) and Vespula Spp. (Yellow Jacket). Toxins 2020, 12, 323. [Google Scholar] [CrossRef]
- Perez-Riverol, A.; Lasa, A.M.; dos Santos-Pinto, J.R.A.; Palma, M.S. Insect Venom Phospholipases A1 and A2: Roles in the Envenoming Process and Allergy. Insect Biochem. Mol. Biol. 2019, 105, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Ebbensgaard, A.; Mordhorst, H.; Overgaard, M.T.; Nielsen, C.G.; Aarestrup, F.M.; Hansen, E.B. Comparative Evaluation of the Antimicrobial Activity of Different Antimicrobial Peptides against a Range of Pathogenic Bacteria. PLoS ONE 2015, 10, e0144611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chernysh, S.; Cociancich, S.; Briand, J.-P.; Hetru, C.; Bulet, P. The Inducible Antibacterial Peptides of the Hemipteran Insect Palomena Prasina: Identification of a Unique Family of Prolinerich Peptides and of a Novel Insect Defensin. J. Insect Physiol. 1996, 42, 81–89. [Google Scholar] [CrossRef]
- Brady, D.; Grapputo, A.; Romoli, O.; Sandrelli, F. Insect Cecropins, Antimicrobial Peptides with Potential Therapeutic Applications. Int. J. Mol. Sci. 2019, 20, 5862. [Google Scholar] [CrossRef] [Green Version]
- Yakovlev, A.Y.; Nesin, A.P.; Simonenko, N.P.; Gordya, N.A.; Tulin, D.V.; Kruglikova, A.A.; Chernysh, S.I. Fat Body and Hemocyte Contribution to the Antimicrobial Peptide Synthesis in Calliphora vicina R.-D. (Diptera: Calliphoridae) Larvae. In Vitro Cell. Dev. Biol. Anim. 2017, 53, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Kuhn-Nentwig, L.; Willems, J.; Seebeck, T.; Shalaby, T.; Kaiser, M.; Nentwig, W. Cupiennin 1a Exhibits a Remarkably Broad, Non-Stereospecific Cytolytic Activity on Bacteria, Protozoan Parasites, Insects, and Human Cancer Cells. Amino Acid 2011, 40, 69–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishii, K.; Hamamoto, H.; Kamimura, M.; Nakamura, Y.; Noda, H.; Imamura, K.; Mita, K.; Sekimizu, K. Insect Cytokine Paralytic Peptide (PP) Induces Cellular and Humoral Immune Responses in the Silkworm Bombyx Mori. J. Biol. Chem. 2010, 285, 28635–28642. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Wang, F.; Dong, Z.; Hua, X.; Xia, Q. Label-Free Quantitative Phosphoproteomic Profiling of Cellular Response Induced by an Insect Cytokine Paralytic Peptide. J. Proteom. 2017, 154, 49–58. [Google Scholar] [CrossRef]
- Shakeel, M.; Xu, X.; Mandal, S.; Jin, F. Role of Serine Protease Inhibitors in Insect-host-pathogen Interactions. Arch. Insect Biochem. Physiol. 2019, 102, e21556. [Google Scholar] [CrossRef]
- Zhu, M.; Hu, X.; Cao, G.; Xue, R.; Gong, C. Functions and Impact of Tal-like Genes in Animals with Regard to Applied Aspects. Appl. Microbiol. Biotechnol. 2018, 102, 6841–6845. [Google Scholar] [CrossRef]
- Pueyo, J.I.; Couso, J.P. Tarsal-Less Peptides Control Notch Signalling through the Shavenbaby Transcription Factor. Dev. Biol. 2011, 355, 183–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, G.; Gong, Y.; Hu, X.; Zhu, M.; Liang, Z.; Huang, L.; Yu, L.; Xu, J.; Li, K.; Zar, M.S.; et al. Identification of Tarsal-Less Peptides from the Silkworm Bombyx Mori. Appl. Microbiol. Biotechnol. 2018, 102, 1809–1822. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Bark, S.; Hook, V.; Bandeira, N. NeuroPedia: Neuropeptide Database and Spectral Library. Bioinformatics 2011, 27, 2772–2773. [Google Scholar] [CrossRef] [Green Version]
- Yeoh, J.G.C.; Pandit, A.A.; Zandawala, M.; Nässel, D.R.; Davies, S.-A.; Dow, J.A.T. DINeR: Database for Insect Neuropeptide Research. Insect Biochem. Mol. Biol. 2017, 86, 9–19. [Google Scholar] [CrossRef]
- Veenstra, J.A. Two Lys-Vasopressin-like Peptides, EFLamide, and Other Phasmid Neuropeptides. Gen. Comp. Endocrinol. 2019, 278, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Zhao, Z. Structure and Function of Neuropeptide F in Insects. J. Integr. Agric. 2020, 19, 1429–1438. [Google Scholar] [CrossRef]
- Nässel, D.R.; Wegener, C. A Comparative Review of Short and Long Neuropeptide F Signaling in Invertebrates: Any Similarities to Vertebrate Neuropeptide Y Signaling? Peptides 2011, 32, 1335–1355. [Google Scholar] [CrossRef]
- Abdoun, K.; Mesnier-Sabin, M.; Baudry-Partiaoglou, N.; Nicolas, P.; Cohen, P. Separation of Oviposition-Stimulating Peptides and Myotropic Factors from Head Extracts of Galleria mellonella L.: Comparative Effects of Myotropic and Non-Myotropic Factors on Egg Laying. J. Comp. Physiol. B 1995, 165, 102–109. [Google Scholar] [CrossRef]
- Wicher, D.; Derst, C.; Gautier, H.; Lapied, B.; Heinemann, S.H.; Agricola, H.J. The satiety signaling neuropeptide perisulfakinin inhibits the activity of central neurons promoting general activity. Front. Cell. Neurosci. 2007, 1, 3. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Cui, D.; Ullah, H.; Hao, K.; Tu, X.; Zhang, Z. Serpin7 Controls Egg Diapause of Migratory Locust (Locusta migratoria) by Regulating Polyphenol Oxidase. FEBS Open Bio 2020, 10, 707–717. [Google Scholar] [CrossRef] [Green Version]
- McClellan-Green, P.; Romano, J.; Oberdörster, E. Does Gender Really Matter in Contaminant Exposure? A Case Study Using Invertebrate Models. Environ. Res. 2007, 104, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Aiello, D.; Siciliano, C.; Mazzotti, F.; Di Donna, L.; Athanassopoulos, C.M.; Napoli, A. A Rapid MALDI MS/MS Based Method for Assessing Saffron (Crocus sativus L.) Adulteration. Food Chem. 2020, 307, 125527. [Google Scholar] [CrossRef] [PubMed]
- Napoli, A.; Athanassopoulos, C.M.; Moschidis, P.; Aiello, D.; Di Donna, L.; Mazzotti, F.; Sindona, G. Solid Phase Isobaric Mass Tag Reagent for Simultaneous Protein Identification and Assay. Anal. Chem. 2010, 82, 5552–5560. [Google Scholar] [CrossRef] [PubMed]
- Naccarato, A.; Furia, E.; Sindona, G.; Tagarelli, A. Multivariate Class Modeling Techniques Applied to Multielement Analysis for the Verification of the Geographical Origin of Chili Pepper. Food Chem. 2016, 206, 217–222. [Google Scholar] [CrossRef] [PubMed]






| IDa | Name a | Sequence | Exact | m/z | |
|---|---|---|---|---|---|
| 1. | FAR14_SARBU | FMRFamide-14 | DPHHDFMRF | 1201.5213 | 1201.54 |
| 2. | NPF1_LEPDE | Neuropeptide NPF-1 | ARGPQLRLRF | 1213.7282 | 1213.75 |
| 3. | ALLTR_ACRHI | Allatotropin-related peptide | GFKNVALSTARGF | 1367.7435 | 1367.77 |
| ALLTR_BANDI | Allatotropin-related peptide | GFKNVALSTARGF | |||
| ALLTR_EUSSE | Allatotropin-related peptide | GFKNVALSTARGF | |||
| ALLTR_NEZVI | Allatotropin-related peptide | GFKNVALSTARGF | |||
| ALLTR_ONCFA | Allatotropin-related peptide | GFKNVALSTARGF | |||
| ALLTR_PENRU | Allatotropin-related peptide | GFKNVALSTARGF | |||
| ALLTR_PERAM | Allatotropin-related peptide | GFKNVALSTARGF | |||
| ALLTR_PYRAP | Allatotropin-related peptide | GFKNVALSTARGF | |||
| 4. | ADFA_TENMO | Antidiuretic factor | VVNTPGHAVSYHVY | 1542.7705 | 1542.80 |
| 5. | TXS6D_CUPSA | Short cationic peptide-6d | INKYREWKNKKN | 1620.8974 | 1620.92 |
| 6. | PPK_SCHGR | Pyrokinin | DGAETPGAAASLWFGPRV—Amide | 1800.9032 | 1800.94 |
| 7. | ALL3_RHOPR | Allatostatin-3 | QVSLKYPEGKMYSFGL | 1846.9413 | 1846.97 |
| 8. | BOL3_BOMPE | Bombolitin-3 | IKIMDILAKLGKVLAHV | 1862.1665 | 1862.19 |
| 9. | BRK_VESMC | Vespulakinin-1 | TATTRRRGRPPGFSPFR | 1960.0741 | 1960.11 |
| 10. | LYC1_LYCSI | M-lycotoxin-Ls3a | GKLQAFLAKMKEIAAQTL | 1961.1257 | 1961.16 |
| 11. | PH1_PERAM | Peptide hormone 1 | SDLTWTYQSPGDPTNSKN | 2010.9045 | 2010.94 |
| 12. | MK2B_PALPR | Metalnikowin-2B | VDKPDYRPRPWPRNMI | 2040.0601 | 2040.10 |
| 13. | LYC40_LYCSI | M-lycotoxin-Ls4a | IASHLAFEKLSKLGSKHTML | 2211.2323 | 2211.28 |
| 14. | PAP2_SPOEX | Paralytic peptide 2 | ENFAGGCTPGYQRTADGRCKPTF | 2476.1138 | 2476.16 |
| 15. | PA11_VESVE | Phospholipase A1 verutoxin-1 (Fragment) | GLLPKVKLVPEQISFILSTRENR | 2637.5455 | 2637.59 |
| 16. | TXC6D_CUPSA | Cupiennin-6d | FINTIKLLIEKYREWKNKQSS | 2638.4721 | 2638.52 |
| 17. | HN423_CYRHA | U3-theraphotoxin-Hhn1r | DCAGYMRECKEKLCCSGYVCSSRWKWCVLPAP | 3671.6222 | 3671.70 |
| 18. | MSPI2_MELSA | Serine protease inhibitor 2 | EISCEPGTTFQDKCNTCRCGKDGKSAAGCTLKACPQ | 3750.6476 | 3750.72 |
| 19. | TXC1C_CUPSA | Cupiennin-1c | GFGSLFKFLAKKVAKTVAKQAAKQGAKYIANKQTE | 3770.1484 | 3770.22 |
| 20. | TALAA_DROME | Peptide tarsal-less AA | LDPTGTYRRPRDTQDSRQKRRQDCLDPTGQY | 3722.8169 | 3722.89 |
| 21. | BX4_LOXGA | Dermonecrotic toxin LgSicTox-beta-LOXN4 | ADSRKPDDRYDMSGNDALGDVKLATYEDNPWETFK | 4019.8357 | 4019.92 |
| 22. | CEC_CALVI | Cecropin | GWLKKIGKKIGRVGQHTRDATIQGLAVAQQAANVAATAR | 4083.3167 | 4083.40 |
| 23. | DIUH1_TENMO | Diuretic hormone 1 | SPTISITAPIDVLRKTWEQERARKQMVKNREFLNSLN | 4369.3566 | 4369.44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aiello, D.; Giglio, A.; Talarico, F.; Vommaro, M.L.; Tagarelli, A.; Napoli, A. Mass Spectrometry-Based Peptide Profiling of Haemolymph from Pterostichus melas Exposed to Pendimethalin Herbicide. Molecules 2022, 27, 4645. https://doi.org/10.3390/molecules27144645
Aiello D, Giglio A, Talarico F, Vommaro ML, Tagarelli A, Napoli A. Mass Spectrometry-Based Peptide Profiling of Haemolymph from Pterostichus melas Exposed to Pendimethalin Herbicide. Molecules. 2022; 27(14):4645. https://doi.org/10.3390/molecules27144645
Chicago/Turabian StyleAiello, Donatella, Anita Giglio, Federica Talarico, Maria Luigia Vommaro, Antonio Tagarelli, and Anna Napoli. 2022. "Mass Spectrometry-Based Peptide Profiling of Haemolymph from Pterostichus melas Exposed to Pendimethalin Herbicide" Molecules 27, no. 14: 4645. https://doi.org/10.3390/molecules27144645
APA StyleAiello, D., Giglio, A., Talarico, F., Vommaro, M. L., Tagarelli, A., & Napoli, A. (2022). Mass Spectrometry-Based Peptide Profiling of Haemolymph from Pterostichus melas Exposed to Pendimethalin Herbicide. Molecules, 27(14), 4645. https://doi.org/10.3390/molecules27144645