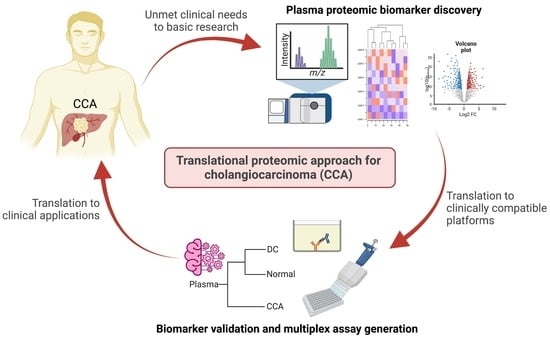
Translational Proteomic Approach for Cholangiocarcinoma Biomarker Discovery, Validation, and Multiplex Assay Development: A Pilot Study
Abstract
:1. Introduction
2. Results
2.1. Discovery of Candidate Biomarkers by Plasma Proteomics
2.2. Rationale for Selection of the Candidate CCA Biomarkers
2.3. Validation of the Candidate Biomarkers by ELISA
2.4. Diagnostic Performance of the Multiplex CCA Markers
3. Discussion
4. Materials and Methods
4.1. Plasma Collection
4.2. Immunodepletion of High Abundance Plasma Proteins
4.3. In-Solution Tryptic Digestion
4.4. Label-Free Quantitation Mass Spectrometry
4.5. ELISA
4.6. Data and Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Laohaviroj, M.; Potriquet, J.; Jia, X.; Suttiprapa, S.; Chamgramol, Y.; Pairojkul, C.; Sithithaworn, P.; Mulvenna, J.; Sripa, B. A comparative proteomic analysis of bile for biomarkers of cholangiocarcinoma. Tumour Biol. 2017, 39, 1010428317705764. [Google Scholar] [CrossRef] [PubMed]
- Tshering, G.; Dorji, P.W.; Chaijaroenkul, W.; Na-Bangchang, K. Biomarkers for the Diagnosis of Cholangiocarcinoma: A Systematic Review. Am. J. Trop. Med. Hyg. 2018, 98, 1788–1797. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Y.; Wei, Y.; Liu, L.M.; Chen, Z.B.; Hu, Q.P.; Pan, S.L. Construction of a model to predict the prognosis of patients with cholangiocarcinoma using alternative splicing events. Oncol. Lett. 2019, 18, 4677–4690. [Google Scholar] [CrossRef] [PubMed]
- Verathamjamras, C.; Weeraphan, C.; Chokchaichamnankit, D.; Watcharatanyatip, K.; Subhasitanont, P.; Diskul-Na-Ayudthaya, P.; Mingkwan, K.; Luevisadpaibul, V.; Chutipongtanate, S.; Champattanachai, V.; et al. Secretomic profiling of cells from hollow fiber bioreactor reveals PSMA3 as a potential cholangiocarcinoma biomarker. Int. J. Oncol. 2017, 51, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Le Faouder, J.; Gigante, E.; Leger, T.; Albuquerque, M.; Beaufrere, A.; Soubrane, O.; Dokmak, S.; Camadro, J.M.; Cros, J.; Paradis, V. Proteomic Landscape of Cholangiocarcinomas Reveals Three Different Subgroups According to Their Localization and the Aspect of Non-Tumor Liver. Proteom. Clin. Appl. 2019, 13, e1800128. [Google Scholar] [CrossRef]
- Aksorn, N.; Roytrakul, S.; Kittisenachai, S.; Leelawat, K.; Chanvorachote, P.; Topanurak, S.; Hamano, S.; Lek-Uthai, U. Novel Potential Biomarkers for Opisthorchis viverrini Infection and Associated Cholangiocarcinoma. In Vivo 2018, 32, 871–878. [Google Scholar] [CrossRef]
- Macias, R.I.R.; Kornek, M.; Rodrigues, P.M.; Paiva, N.A.; Castro, R.E.; Urban, S.; Pereira, S.P.; Cadamuro, M.; Rupp, C.; Loosen, S.H.; et al. Diagnostic and prognostic biomarkers in cholangiocarcinoma. Liver Int. 2019, 39, 108–122. [Google Scholar] [CrossRef]
- Marrero, J.A. Biomarkers in cholangiocarcinoma. Clin. Liver Dis. 2014, 3, 101–103. [Google Scholar] [CrossRef]
- Loosen, S.H.; Vucur, M.; Trautwein, C.; Roderburg, C.; Luedde, T. Circulating Biomarkers for Cholangiocarcinoma. Dig. Dis. 2018, 36, 281–288. [Google Scholar] [CrossRef]
- Macias, R.I.R.; Banales, J.M.; Sangro, B.; Muntane, J.; Avila, M.A.; Lozano, E.; Perugorria, M.J.; Padillo, F.J.; Bujanda, L.; Marin, J.J.G. The search for novel diagnostic and prognostic biomarkers in cholangiocarcinoma. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1468–1477. [Google Scholar] [CrossRef]
- Silsirivanit, A.; Sawanyawisuth, K.; Riggins, G.J.; Wongkham, C. Cancer biomarker discovery for cholangiocarcinoma: The high-throughput approaches. J. Hepatobiliary Pancreat Sci. 2014, 21, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Blechacz, B.; Gores, G.J. Cholangiocarcinoma: Advances in pathogenesis, diagnosis, and treatment. Hepatology 2008, 48, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Van Beers, B.E. Diagnosis of cholangiocarcinoma. HPB 2008, 10, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, D.J.; Chen, J.; Liu, W.; Li, J.W.; Jiang, P.; Zhao, X.; Guo, F.; Li, X.W.; Wang, S.G. Application of Joint Detection of AFP, CA19-9, CA125 and CEA in Identification and Diagnosis of Cholangiocarcinoma. Asian Pac. J. Cancer Prev. 2015, 16, 3451–3455. [Google Scholar] [CrossRef]
- Ince, A.T.; Yildiz, K.; Baysal, B.; Danalioglu, A.; Kocaman, O.; Tozlu, M.; Gangarapu, V.; Sarbay Kemik, A.; Uysal, O.; Senturk, H. Roles of serum and biliary CEA, CA19-9, VEGFR3, and TAC in differentiating between malignant and benign biliary obstructions. Turk. J. Gastroenterol. 2014, 25, 162–169. [Google Scholar] [CrossRef]
- Shigehara, K.; Yokomuro, S.; Ishibashi, O.; Mizuguchi, Y.; Arima, Y.; Kawahigashi, Y.; Kanda, T.; Akagi, I.; Tajiri, T.; Yoshida, H.; et al. Real-time PCR-based analysis of the human bile microRNAome identifies miR-9 as a potential diagnostic biomarker for biliary tract cancer. PLoS ONE 2011, 6, e23584. [Google Scholar] [CrossRef] [PubMed]
- Takenami, T.; Maeda, S.; Karasawa, H.; Suzuki, T.; Furukawa, T.; Morikawa, T.; Takadate, T.; Hayashi, H.; Nakagawa, K.; Motoi, F.; et al. Novel biomarkers distinguishing pancreatic head Cancer from distal cholangiocarcinoma based on proteomic analysis. BMC Cancer 2019, 19, 318. [Google Scholar] [CrossRef] [PubMed]
- Padden, J.; Megger, D.A.; Bracht, T.; Reis, H.; Ahrens, M.; Kohl, M.; Eisenacher, M.; Schlaak, J.F.; Canbay, A.E.; Weber, F.; et al. Identification of novel biomarker candidates for the immunohistochemical diagnosis of cholangiocellular carcinoma. Mol. Cell. Proteom. 2014, 13, 2661–2672. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.G.; Yoon, S.Y.; Oh, J.H.; Jeon, Y.J.; Kim, M.; Kim, J.M.; Byun, S.S.; Yang, J.O.; Kim, J.H.; Kim, D.G.; et al. Identification of intrahepatic cholangiocarcinoma related genes by comparison with normal liver tissues using expressed sequence tags. Biochem. Biophys. Res. Commun. 2006, 345, 1022–1032. [Google Scholar] [CrossRef]
- Darby, I.A.; Vuillier-Devillers, K.; Pinault, E.; Sarrazy, V.; Lepreux, S.; Balabaud, C.; Bioulac-Sage, P.; Desmouliere, A. Proteomic analysis of differentially expressed proteins in peripheral cholangiocarcinoma. Cancer Microenviron. 2010, 4, 73–91. [Google Scholar] [CrossRef] [Green Version]
- Srisomsap, C.; Sawangareetrakul, P.; Subhasitanont, P.; Chokchaichamnankit, D.; Chiablaem, K.; Bhudhisawasdi, V.; Wongkham, S.; Svasti, J. Proteomic studies of cholangiocarcinoma and hepatocellular carcinoma cell secretomes. J. Biomed. Biotechnol. 2010, 2010, 437143. [Google Scholar] [CrossRef]
- Kimawaha, P.; Jusakul, A.; Junsawang, P.; Thanan, R.; Titapun, A.; Khuntikeo, N.; Techasen, A. Establishment of a Potential Serum Biomarker Panel for the Diagnosis and Prognosis of Cholangiocarcinoma Using Decision Tree Algorithms. Diagnostics 2021, 11, 589. [Google Scholar] [CrossRef] [PubMed]
- Duangkumpha, K.; Stoll, T.; Phetcharaburanin, J.; Yongvanit, P.; Thanan, R.; Techasen, A.; Namwat, N.; Khuntikeo, N.; Chamadol, N.; Roytrakul, S.; et al. Discovery and Qualification of Serum Protein Biomarker Candidates for Cholangiocarcinoma Diagnosis. J. Proteome. Res. 2019, 18, 3305–3316. [Google Scholar] [CrossRef]
- Puetkasichonpasutha, J.; Namwat, N.; Sa-Ngiamwibool, P.; Titapun, A.; Suthiphongchai, T. Evaluation of p53 and Its Target Gene Expression as Potential Biomarkers of Cholangiocarcinoma in Thai Patients. Asian Pac. J. Cancer Prev. 2020, 21, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Deng, X.; Zhan, Q.; Shen, B.; Jin, X.; Zhu, Z.; Chen, H.; Li, H.; Peng, C. A prospective proteomic-based study for identifying potential biomarkers for the diagnosis of cholangiocarcinoma. J. Gastrointest. Surg. 2013, 17, 1584–1591. [Google Scholar] [CrossRef] [PubMed]
- Changbumrung, S.; Migasena, P.; Supawan, V.; Juttijudata, P.; Buavatana, T. Serum protease inhibitors in opisthorchiasis, hepatoma, cholangiocarcinoma, and other liver diseases. Southeast Asian J. Trop. Med. Public Health 1988, 19, 299–305. [Google Scholar]
- Changbumrung, S.; Migasena, P.; Supawan, V.; Buavatana, T.; Migasena, S. Alpha 1-antitrypsin, alpha 1-antichymotrypsin and alpha 2-macroglobulin in human liver fluke (opisthorchiasis). Trop. Parasitol. 1982, 33, 195–197. [Google Scholar]
- Chang, T.T.; Ho, C.H. Plasma proteome atlas for differentiating tumor stage and post-surgical prognosis of hepatocellular carcinoma and cholangiocarcinoma. PLoS ONE 2020, 15, e0238251. [Google Scholar] [CrossRef] [PubMed]
- Tolek, A.; Wongkham, C.; Proungvitaya, S.; Silsirivanit, A.; Roytrakul, S.; Khuntikeo, N.; Wongkham, S. Serum alpha1beta-glycoprotein and afamin ratio as potential diagnostic and prognostic markers in cholangiocarcinoma. Exp. Biol. Med. 2012, 237, 1142–1149. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, X.; Maglic, D.; Dill, M.T.; Mojumdar, K.; Ng, P.K.; Jeong, K.J.; Tsang, Y.H.; Moreno, D.; Bhavana, V.H.; et al. Comprehensive Molecular Characterization of the Hippo Signaling Pathway in Cancer. Cell Rep. 2018, 25, 1304–1317. [Google Scholar] [CrossRef]
- Nair, A.; Ingram, N.; Verghese, E.T.; Wijetunga, I.; Markham, A.F.; Wyatt, J.; Prasad, K.R.; Coletta, P.L. Neutrophil Gelatinase-associated Lipocalin as a Theragnostic Marker in Perihilar Cholangiocarcinoma. Anticancer Res. 2018, 38, 6737–6744. [Google Scholar] [CrossRef] [PubMed]
- Leelawat, K.; Narong, S.; Wannaprasert, J.; Leelawat, S. Serum NGAL to Clinically Distinguish Cholangiocarcinoma from Benign Biliary Tract Diseases. Int. J. Hepatol. 2011, 2011, 873548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chutipongtanate, S.; Chatchen, S.; Svasti, J. Plasma prefractionation methods for proteomic analysis and perspectives in clinical applications. Proteom. Clin. Appl. 2017, 11, 1600135. [Google Scholar] [CrossRef] [PubMed]
- Mattsson-Carlgren, N.; Palmqvist, S.; Blennow, K.; Hansson, O. Publisher Correction: Increasing the reproducibility of fluid biomarker studies in neurodegenerative studies. Nat. Commun. 2021, 12, 196. [Google Scholar] [CrossRef]
- Geyer, P.E.; Holdt, L.M.; Teupser, D.; Mann, M. Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol. 2017, 13, 942. [Google Scholar] [CrossRef]
- Sašo Džeroski, P.P.B.Ž. Machine Learning, Ensemble Methods in. In Encyclopedia of Complexity and Systems Science; Meyers, R.A., Ed.; Springer: New York, NY, USA, 2009; pp. 5317–5325. [Google Scholar]
- Dwivedi, P.; Chutipongtanate, S.; Muench, D.E.; Azam, M.; Grimes, H.L.; Greis, K.D. SWATH-Proteomics of Ibrutinib’s Action in Myeloid Leukemia Initiating Mutated G-CSFR Signaling. Proteom. Clin. Appl. 2020, 14, e1900144. [Google Scholar] [CrossRef]





| Pooled Sample | Gender | Age | Condition/Disease | CCA Stage |
|---|---|---|---|---|
| M | 46 | Cholangiocarcinoma, perihilar | I | |
| pCCA1 | M | 51 | Cholangiocarcinoma, distal | IIa |
| M | 73 | Cholangiocarcinoma, distal | IIb | |
| M | 67 | Cholangiocarcinoma, intrahepatic | III | |
| pCCA2 | F | 55 | Cholangiocarcinoma, intrahepatic | IIIA |
| F | 46 | Cholangiocarcinoma, intrahepatic | IIIA | |
| M | 50 | Cholangiocarcinoma, metastasis | IV | |
| pCCA3 | M | 55 | Cholangiocarcinoma, intrahepatic | IV |
| F | 51 | Cholangiocarcinoma, intrahepatic | IV | |
| F | 51 | Healthy | - | |
| pN1 | M | 56 | Healthy | - |
| F | 52 | Healthy | - | |
| M | 55 | Healthy | - | |
| pN2 | F | 59 | Healthy | - |
| F | 50 | Healthy | - | |
| M | 54 | Healthy | - | |
| pN3 | F | 56 | Healthy | - |
| M | 66 | Healthy | - | |
| F | 72 | HCC, chronic cholecystitis | - | |
| pDC1 | M | 52 | HCC, cirrhosis | - |
| F | 61 | HCC | - | |
| M | 34 | Chronic HBV infection | - | |
| pDC2 | F | 64 | Chronic cholecystitis, DM, HT | - |
| M | 56 | Periductal fibrosis | - | |
| F | 33 | Focal nodular hyperplasia, liver | - | |
| pDC3 | M | 59 | Granulomatous inflammation, CBD | - |
| F | 64 | Gastrointestinal stromal tumor | - |
| Gene Name | Accession | Protein Name | Rationale for Selection | Reference |
|---|---|---|---|---|
| S100A9 | S10A9_HUMAN | Protein S100-A9 | Significantly upregulated in pCCA vs. pN and pDC (p < 0.001) Previously identified as a CCA biomarker in multiple independent studies | This study [22,23,24,25] |
| SERPINA3 | AACT_HUMAN | Alpha-1-antichymotrypsin | Significantly upregulated in pCCA vs. pN and pDC (p < 0.001) Previously proposed as a candidate biomarker of opisthorchiasis-associated CCA | This study [26,27] |
| AFM | AFAM_HUMAN | Afamin | Significantly downregulated in pCCA vs. pN (p < 0.001) Previously identified as a biomarker of advanced CCA with poor prognosis | This study [28,29] |
| TAOK3 | TAOK3_HUMAN | Serine/threonine-protein kinase TAO3 | Significantly upregulated in pCCA vs. pDC (p < 0.001) A tumor suppressor gene with genomic evidence of significant alteration in CCA | This study [30] |
| NGAL | NGAL_HUMAN | Neutrophil gelatinase-associated lipocalin | Previously identified as a biomarker of perihilar CCA, which could distinguish CCA from benign biliary tract diseases | [21,31,32] |
| PSMA3 | PSA3_HUMAN | Proteasome subunit alpha type 3 | Previously identified as a CCA biomarker from the CCA cell secretome and successfully validated using an antibody-based assay in 12 clinical plasma samples (5 normal, 4 CCA, 3 DC) | [4] a |
| AMBP | AMBP_HUMAN | Alpha-1 microglobulin | Previously identified as a biomarker of intrahepatic CCA | [19,20] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watcharatanyatip, K.; Chutipongtanate, S.; Chokchaichamnankit, D.; Weeraphan, C.; Mingkwan, K.; Luevisadpibul, V.; Newburg, D.S.; Morrow, A.L.; Svasti, J.; Srisomsap, C. Translational Proteomic Approach for Cholangiocarcinoma Biomarker Discovery, Validation, and Multiplex Assay Development: A Pilot Study. Molecules 2022, 27, 5904. https://doi.org/10.3390/molecules27185904
Watcharatanyatip K, Chutipongtanate S, Chokchaichamnankit D, Weeraphan C, Mingkwan K, Luevisadpibul V, Newburg DS, Morrow AL, Svasti J, Srisomsap C. Translational Proteomic Approach for Cholangiocarcinoma Biomarker Discovery, Validation, and Multiplex Assay Development: A Pilot Study. Molecules. 2022; 27(18):5904. https://doi.org/10.3390/molecules27185904
Chicago/Turabian StyleWatcharatanyatip, Kamolwan, Somchai Chutipongtanate, Daranee Chokchaichamnankit, Churat Weeraphan, Kanokwan Mingkwan, Virat Luevisadpibul, David S. Newburg, Ardythe L. Morrow, Jisnuson Svasti, and Chantragan Srisomsap. 2022. "Translational Proteomic Approach for Cholangiocarcinoma Biomarker Discovery, Validation, and Multiplex Assay Development: A Pilot Study" Molecules 27, no. 18: 5904. https://doi.org/10.3390/molecules27185904
APA StyleWatcharatanyatip, K., Chutipongtanate, S., Chokchaichamnankit, D., Weeraphan, C., Mingkwan, K., Luevisadpibul, V., Newburg, D. S., Morrow, A. L., Svasti, J., & Srisomsap, C. (2022). Translational Proteomic Approach for Cholangiocarcinoma Biomarker Discovery, Validation, and Multiplex Assay Development: A Pilot Study. Molecules, 27(18), 5904. https://doi.org/10.3390/molecules27185904