Identification of SARS-CoV-2 Main Protease Inhibitors from a Library of Minor Cannabinoids by Biochemical Inhibition Assay and Surface Plasmon Resonance Characterized Binding Affinity
Abstract
:1. Introduction
2. Results
2.1. Inhibitory Effects of the THC-Type Cannabinoids on SARS-CoV-2 Mpro
2.2. Inhibitory Effects of the CBG-Type Cannabinoids on SARS-CoV-2 Mpro
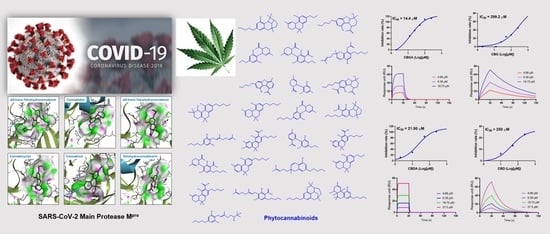
2.3. Inhibitory Effects of the CBD-Type Cannabinoids on SARS-CoV-2 Mpro
2.4. Inhibitory Effects of CBC-Type Cannabinoids on SARS-CoV-2 Mpro
2.5. Inhibitory Effects of Other Cannabinoids on SARS-CoV-2 Mpro
2.6. Validation of the SPR Method to Measure the Binding Affinity with SARS-CoV-2 Mpro Protein
2.7. Decarboxylation of CBGA and CBDA Attenuates the Anti-Mpro Activity
3. Discussion
4. Materials and Methods
4.1. Minor Cannabinoids
4.2. Enzyme Inhibition Assay
4.3. Surface Plasmon Resonance
4.4. Molecular Modeling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Li, H.; Liu, S.M.; Yu, X.H.; Tang, S.L.; Tang, C.K. Coronavirus disease 2019 (COVID-19): Current status and future perspectives. Int. J. Antimicrob. Agents 2020, 55, 105951. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.; Ni, Z.; Hu, Y.; Liang, W.; Ou, C.; He, J.; Liu, L.; Shan, H.; Lei, C.; Hui, D.S.C.; et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Hu, S.; Gao, J. Discovering drugs to treat coronavirus disease 2019 (COVID-19). Drug Discov. Ther. 2020, 14, 58–60. [Google Scholar] [CrossRef]
- Kitazato, K.; Wang, Y.; Kobayashi, N. Viral infectious disease and natural products with antiviral activity. Drug Discov. Ther. 2007, 1, 14–22. [Google Scholar]
- El Sayed, K.A. Natural products as antiviral agents. Stud. Nat. Prod. Chem. 2000, 24, 473. [Google Scholar] [CrossRef]
- Islam, M.T.; Sarkar, C.; El-Kersh, D.M.; Jamaddar, S.; Uddin, S.J.; Shilpi, J.A.; Mubarak, M.S. Natural products and their derivatives against coronavirus: A review of the non-clinical and pre-clinical data. Phytother. Res. 2020, 34, 2471–2492. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Park, R.; Jang, M.; Park, Y.I.; Park, Y. Coronavirus enzyme inhibitors-experimentally proven natural compounds from plants. J. Microbiol. 2022, 60, 347–354. [Google Scholar] [CrossRef]
- Khoury, M.; Rocco, P.R.M.; Phinney, D.G.; Krampera, M.; Martin, I.; Viswanathan, S.; Nolta, J.A.; LeBlanc, K.; Galipeau, J.; Weiss, D.J. Cell-based therapies for coronavirus disease 2019: Proper clinical investigations are essential. Cytotherapy 2020, 22, 602–605. [Google Scholar] [CrossRef]
- Yuan, L.; Tang, Q.; Cheng, T.; Xia, N. Animal models for emerging coronavirus: Progress and new insights. Emerg. Microbes Infect. 2020, 9, 949–961. [Google Scholar] [CrossRef]
- Narkhede, R.R.; Pise, A.V.; Cheke, R.S.; Shinde, S.D. Recognition of natural products as potential inhibitors of COVID-19 main protease (Mpro): In-silico evidences. Nat. Prod. Bioprospect. 2020, 10, 297–306. [Google Scholar] [CrossRef]
- Orhan, I.E.; Senol Deniz, F.S. Natural products as potential leads against coronaviruses: Could they be encouraging structural models against SARS-CoV-2? Nat. Prod. Bioprospect. 2020, 10, 171–186. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, S.; Nitsche, C. The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020, 30, 127377. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef]
- Saied, E.M.; El-Maradny, Y.A.; Osman, A.A.; Darwish, A.M.G.; Nahas, H.H.A.; Niedbała, G.; Piekutowska, M.; Abdel-Rahman, M.A.; Balbool, B.A.; Abdel-Azeem, A.M. A comprehensive review about the molecular structure of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): Insights into natural products against COVID-19. Pharmaceutics 2021, 13, 1759. [Google Scholar] [CrossRef]
- Adem, S.; Eyupoglu, V.; Sarfraz, I.; Rasul, A.; Ali, M. Identification of potent COVID-19 main protease (Mpro) inhibitors from natural polyphenols: An in silico strategy unveils a hope against CORONA. Preprints 2020. [Google Scholar] [CrossRef]
- Khan, M.T.; Ali, A.; Wang, Q.; Irfan, M.; Khan, A.; Zeb, M.T.; Zhang, Y.J.; Chinnasamy, S.; Wei, D.Q. Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2—A molecular dynamic study. J. Biomol. Struct. Dyn. 2020, 39, 3627–3637. [Google Scholar] [CrossRef]
- Tallei, T.E.; Tumilaar, S.G.; Niode, N.J.; Fatimawali, F.; Kepel, B.J.; Idroes, R.; Effendi, Y. Potential of plant bioactive compounds as SARS-CoV-2 main protease (Mpro) and spike (S) glycoprotein inhibitors: A molecular docking study. Preprints 2020. [Google Scholar] [CrossRef]
- Kumar, A.; Choudhir, G.; Shukla, S.K.; Sharma, M.; Tyagi, P.; Bhushan, A.; Rathore, M. Identification of phytochemical inhibitors against main protease of COVID-19 using molecular modeling approaches. J. Biomol. Struct. Dyn. 2020, 39, 3760–3770. [Google Scholar] [CrossRef]
- Tagne, A.M.; Pacchetti, B.; Sodergren, M.; Sodergren, M.; Cosentino, M.; Marino, F. Cannabidiol for viral diseases: Hype or hope? Cannabis Cannabinoid Res. 2020, 5, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Hill, K.P.; Hill, K.P. Cannabinoids and the coronavirus. Cannabis Cannabinoid Res. 2020, 5, 118–120. [Google Scholar] [CrossRef] [PubMed]
- Raj, V.; Park, J.G.; Cho, K.H.; Choi, P.; Kim, T.; Ham, J.; Lee, J. Assessment of antiviral potencies of cannabinoids against SARS-CoV-2 using computational and in vitro approaches. Int. J. Biol. Macromol. 2021, 168, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Altyar, A.E.; Youssef, F.S.; Kurdi, M.M.; Bifari, R.J.; Ashour, M.L. The role of Cannabis Sativa, L. as a source of cannabinoids against Coronavirus 2 (SARS-CoV-2): An in silico study to evaluate their activities and ADMET properties. Molecules 2022, 27, 2797. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Li, H.; Liu, C.; Seeram, N.P. Evaluation of cannabidiol’s inhibitory effect on alpha-glucosidase and its stability in simulated gastric and intestinal fluids. J. Cannabis Res. 2021, 3, 20. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, H.; Xu, F.; Jiang, X.; Ma, H.; Seeram, N.P. Cannabidiol protects human skin keratinocytes from hydrogen-peroxide-induced oxidative stress via modulation of the caspase-1-IL-1β axis. J. Nat. Prod. 2021, 84, 1563–1572. [Google Scholar] [CrossRef] [PubMed]
- Puopolo, T.; Liu, C.; Ma, H.; Seeram, N.P. Inhibitory effects of cannabinoids on acetylcholinesterase and butyrylcholinesterase enzyme activities. Med. Cannabis Cannabinoids 2022, 5, 85–94. [Google Scholar] [CrossRef]
- Van Breemen, R.B.; Muchiri, R.N.; Bates, T.A.; Weinstein, J.B.; Leier, H.C.; Farley, S.; Tafesse, F.G. Cannabinoids block cellular entry of SARS-CoV-2 and the emerging variants. J. Nat. Prod. 2022, 85, 176–184. [Google Scholar] [CrossRef]
- Wang, B.; Kovalchuk, A.; Li, D.; Ilnytskyy, Y.; Kovalchuk, I. In Search of Preventative Strategies: Novel anti- inflammatory high-CBD Cannabis sativa extracts modulate ACE2 expression in COVID-19 gateway tissues. Preprint 2020, 1–12. [Google Scholar] [CrossRef]
- Sun, H.; Li, Y.; Zhang, X.; Lei, Y.; Ding, W.; Zhao, X.; Wang, H.; Song, X.; Yao, Q.; Zhang, Y.; et al. Synthesis, α-glucosidase inhibitory and molecular docking studies of prenylated and geranylated flavones, isoflavones and chalcones. Bioorg. Med. Chem. Lett. 2015, 25, 4567–4571. [Google Scholar] [CrossRef]
- Chen, X.; Mukwaya, E.; Wong, M.S.; Zhang, Y. A systematic review on biological activities of prenylated flavonoids. Pharm. Biol. 2014, 52, 655–660. [Google Scholar] [CrossRef]
- Li, H.; Xu, F.; Liu, C.; Cai, A.; Dain, J.A.; Li, D.; Seeram, N.P.; Cho, B.P.; Ma, H. Inhibitory effects and surface plasmon resonance-based binding affinities of dietary hydrolyzable tannins and their gut microbial metabolites on SARS-CoV-2 main protease. J. Agric. Food Chem. 2021, 69, 12197–12208. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Cai, A.; Li, H.; Deng, N.; Cho, B.P.; Seeram, N.P.; Ma, H. Characterization of molecular interactions between cannabidiol and human plasma proteins (serum albumin and γ-globulin) by surface plasmon resonance, microcalorimetry, and molecular docking. J. Pharm. Biomed. Anal. 2022, 214, 114750. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xie, W.; Xue, X.; Yang, K.; Ma, J.; Liang, W.; Zhao, Q.; Zhou, Z.; Pei, D.; Ziebuhr, J.; et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005, 3, e324. [Google Scholar] [CrossRef]











| Ligand | Ka (1/Ms) | Kd (1/s) | KD (M) |
|---|---|---|---|
| Mpro 11a | 7715 | 0.1464 | 1.90 × 10−5 |
| Mpro 11b | 7975 | 0.05533 | 6.94 × 10−6 |
| Mpro N3 | 6631 | 0.02898 | 4.37 × 10−6 |
| Ligand | Ka (1/Ms) | Kd (1/s) | KD (M) |
|---|---|---|---|
| CBGA | 389.8 | 0.01516 | 3.89 × 10−5 |
| CBG | 12160 | 1.104 | 9.01 × 10−5 |
| Predicted Binding Energies | CBGA (kcal/mol) | CBG (kcal/mol) |
|---|---|---|
| Free binding energy | −4.61 | −4.85 |
| Intermolecular energy | −8.48 | −8.13 |
| Total internal energy | −2.29 | −1.27 |
| Torsional free energy | 3.88 | 3.28 |
| Ligand | Ka (1/Ms) | Kd (1/s) | KD (M) |
|---|---|---|---|
| CBDA | 12160 | 1.104 | 9.08 × 10−5 |
| CBD | 533.3 | 0.04173 | 7.83 × 10−5 |
| Predicted Binding Energies | CBDA (kcal/mol) | CBD (kcal/mol) |
|---|---|---|
| Free binding energy | −6.51 | −6.85 |
| Intermolecular energy | −9.49 | −9.24 |
| Total internal energy | −2.39 | −1.29 |
| Torsional free energy | 2.98 | 2.39 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Puopolo, T.; Li, H.; Cai, A.; Seeram, N.P.; Ma, H. Identification of SARS-CoV-2 Main Protease Inhibitors from a Library of Minor Cannabinoids by Biochemical Inhibition Assay and Surface Plasmon Resonance Characterized Binding Affinity. Molecules 2022, 27, 6127. https://doi.org/10.3390/molecules27186127
Liu C, Puopolo T, Li H, Cai A, Seeram NP, Ma H. Identification of SARS-CoV-2 Main Protease Inhibitors from a Library of Minor Cannabinoids by Biochemical Inhibition Assay and Surface Plasmon Resonance Characterized Binding Affinity. Molecules. 2022; 27(18):6127. https://doi.org/10.3390/molecules27186127
Chicago/Turabian StyleLiu, Chang, Tess Puopolo, Huifang Li, Ang Cai, Navindra P. Seeram, and Hang Ma. 2022. "Identification of SARS-CoV-2 Main Protease Inhibitors from a Library of Minor Cannabinoids by Biochemical Inhibition Assay and Surface Plasmon Resonance Characterized Binding Affinity" Molecules 27, no. 18: 6127. https://doi.org/10.3390/molecules27186127
APA StyleLiu, C., Puopolo, T., Li, H., Cai, A., Seeram, N. P., & Ma, H. (2022). Identification of SARS-CoV-2 Main Protease Inhibitors from a Library of Minor Cannabinoids by Biochemical Inhibition Assay and Surface Plasmon Resonance Characterized Binding Affinity. Molecules, 27(18), 6127. https://doi.org/10.3390/molecules27186127