Essential Oil Chemotypes and Genetic Variability of Cinnamomum verum Leaf Samples Commercialized and Cultivated in the Amazon
Abstract
:1. Introduction
2. Results and Discussion
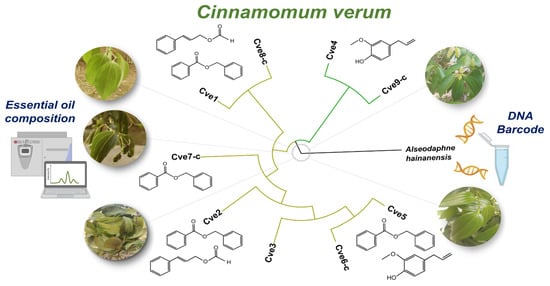
2.1. Chemical Composition and Multivariate Analysis
| Oil Yield (%) | 0.54 | 1.67 | 1.30 | 1.90 | 2.50 | 0.70 | 0.50 | 0.80 | 2.50 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Constituent (%) | IR(L) | IR(C) | CVe1 | CVe2 | CVe3 | CVe4 | CVe5 | CVe6-c | CVe7-c | CVe8-c | CVe9-c |
| Ethylbenzene | 857 1 | 859 | 0.01 | ||||||||
| Styrene | 891 1 | 891 | 0.07 | 0.01 | 0.02 | 0.04 | 0.01 | 0.01 | 0.01 | ||
| Tricyclene | 921 2 | 922 | 0.02 | 0.01 | 0.01 | 0.01 | |||||
| α-Thujene | 924 2 | 925 | 0.06 | 0.02 | 0.01 | 0.06 | 0.06 | 0.03 | |||
| α-Pinene | 932 2 | 933 | 2.89 | 1.18 | 1.52 | 0.23 | 0.12 | 1.69 | 1.51 | 0.33 | 0.23 |
| Camphene | 946 2 | 947 | 1.45 | 0.6 | 1.03 | 0.11 | 0.07 | 0.84 | 0.82 | 0.12 | 0.13 |
| 6-Methylheptan-2-ol | 958 2 | 950 | 0.14 | ||||||||
| Benzaldehyde | 952 2 | 957 | 2.36 | 0.51 | 2.13 | 0.11 | 0.15 | 0.45 | 0.8 | 0.44 | 0.26 |
| Sabinene | 969 2 | 972 | 0.04 | 0.02 | 0.01 | 0.03 | 0.08 | 0.05 | 0.01 | 0.02 | |
| β-Pinene | 974 2 | 976 | 1.36 | 0.59 | 0.87 | 0.1 | 0.1 | 0.76 | 0.85 | 0.15 | 0.15 |
| Myrcene | 988 2 | 989 | 0.28 | 0.17 | 0.07 | 0.01 | 0.02 | 0.22 | 0.3 | 0.06 | 0.02 |
| Mesitylene | 994 2 | 993 | 0.01 | ||||||||
| n-Decane | 1000 2 | 998 | 0.01 | 0.02 | 0.02 | 0.01 | |||||
| α-Phellandrene | 1004 2 | 1004 | 0.48 | 0.25 | 0.03 | 0.07 | 0.03 | 0.17 | 0.74 | 0.57 | |
| δ-3-Carene | 1008 2 | 1010 | 0.04 | 0.03 | 0.01 | 0.03 | 0.03 | ||||
| α-Terpinene | 1014 2 | 1016 | 0.02 | 0.04 | 0.03 | 0.3 | 0.15 | 0.01 | 0.03 | ||
| p-Cymene | 1020 2 | 1023 | 0.34 | 0.1 | 0.53 | 0.04 | 0.06 | 0.11 | 0.28 | 0.26 | 0.02 |
| β-Phellandrene | 1025 2 | 1027 | 0.29 | 1.45 | 0.32 | ||||||
| Limonene | 1024 2 | 1027 | 0.96 | 0.66 | 0.59 | 0.07 | 1.34 | 0.18 | |||
| 1,8-Cineole | 1026 2 | 1030 | 0.01 | ||||||||
| Benzyl alcohol | 1026 2 | 1031 | 1.06 | 0.82 | 0.71 | 0.06 | 0.63 | 0.67 | 0.26 | 0.03 | |
| (Z)-β-Ocimene | 1032 2 | 1035 | 0.01 | 0.1 | 0.04 | 0.05 | |||||
| Butyl 2-methylbutyrate | 1042 1 | 1038 | 0.02 | ||||||||
| Salicylaldehyde | 1039 2 | 1040 | 0.03 | 0.03 | |||||||
| (E)-β-Ocimene | 1044 2 | 1045 | 0.02 | 0.07 | 0.13 | 0.89 | 0.01 | ||||
| γ-Terpinene | 1054 2 | 1056 | 0.02 | 0.05 | 0.04 | 0.01 | |||||
| Acetophenone | 1059 2 | 1063 | 0.04 | ||||||||
| cis-Linalool oxide (furanoid) | 1067 2 | 1070 | 0.01 | 0.02 | 0.01 | ||||||
| trans-Linalool oxide (furanoid) | 1084 2 | 1087 | 0.04 | ||||||||
| Terpinolene | 1086 2 | 1087 | 0.09 | 0.08 | 0.09 | 0.12 | 0.06 | ||||
| Methyl benzoate | 1088 2 | 1094 | 0.01 | 0.02 | |||||||
| Linalool | 1095 2 | 1099 | 1.97 | 3.66 | 0.05 | 0.54 | 3.13 | 3.69 | 0.86 | 2.26 | |
| 2-Methylbutyl 2-methylbutyrate | 1100 2 | 1103 | 0.04 | 0.03 | 0.02 | 0.09 | 0.02 | 0.02 | |||
| α-Campholenal | 1122 2 | 1124 | 0.09 | ||||||||
| (trans)-p-Menth-2-en-1ol | 1136 2 | 1137 | 0.01 | 0.01 | |||||||
| (trans)-Pinocarveol | 1135 2 | 1137 | 0.16 | ||||||||
| Camphor | 1141 2 | 1143 | 0.03 | 0.03 | 0.04 | 0.01 | 0.04 | ||||
| Pinocarvone | 1160 2 | 1161 | 0.36 | ||||||||
| Hydrocinnamaldehyde | 1599 2 | 1160 | 1.4 | 0.16 | 0.01 | 0.18 | 0.08 | 0.2 | 0.45 | ||
| Benzyl acetate | 1157 2 | 1162 | 0.72 | 0.12 | 0.03 | 0.75 | 0.04 | 0.1 | 0.04 | ||
| Borneol | 1165 2 | 1164 | 0.26 | 0.14 | 0.22 | 0.03 | 0.11 | 0.16 | 0.01 | 0.05 | |
| Pyruvophenone | 1169 1 | 1165 | 0.14 | ||||||||
| Ethyl benzoate | 1169 2 | 1169 | 0.11 | 0.06 | 0.04 | 0.05 | 0.04 | 0.03 | |||
| Terpinen-4-ol | 1174 2 | 1176 | 0.1 | 0.09 | 0.09 | 0.05 | 0.1 | 0.14 | 0.04 | 0.04 | |
| Naphthalene | 1178 2 | 1181 | 0.04 | ||||||||
| p-Cymen-8-ol | 1179 2 | 1183 | 0.02 | ||||||||
| α-Terpineol | 1186 2 | 1189 | 0.34 | 0.32 | 0.3 | 0.03 | 0.05 | 0.18 | 0.43 | 0.15 | 0.11 |
| (4Z)-Decenal | 1193 2 | 1192 | 0.08 | 0.01 | |||||||
| Myrtenol | 1194 2 | 1195 | 0.03 | 0.12 | |||||||
| Methyl chavicol | 1195 2 | 1197 | 0.01 | ||||||||
| (Z)-Cinnamaldehyde | 1217 2 | 1217 | 0.17 | 0.03 | 0.06 | ||||||
| Hydrocinnamyl alcohol | 1124 2 | 1228 | 0.13 | 0.22 | 0.02 | 0.05 | |||||
| Chavicol | 1247 2 | 1253 | 0.03 | 0.11 | 0.05 | 0.06 | |||||
| 2-Phenylethyl acetate | 1254 2 | 1255 | 0.05 | 0.03 | |||||||
| (E)-Cinnamaldehyde | 1267 2 | 1274 | 19.74 | 2.4 | 3.51 | 1 | 3.03 | 3.04 | 3.76 | 10.82 | 4.86 |
| (E)-Cinnamyl alcohol | 1303 2 | 1303 | 0.25 | 1.27 | 0.04 | ||||||
| Isobutyl benzoate | 1327 2 | 1327 | 0.01 | 0.04 | 0.01 | ||||||
| δ-Elemene | 1335 2 | 1337 | 0.07 | 0.04 | 0.19 | 0.06 | |||||
| α-Cubebene | 1345 2 | 1349 | 0.09 | 0.04 | 0.04 | ||||||
| Eugenol | 1356 2 | 1356 | 1.07 | 2.13 | 0.7 | 91 | 90.15 | 54.51 | 1.32 | 1.69 | 85.68 |
| Hydrocinnamyl acetate | 1366 2 | 1370 | 2.91 | 0.29 | 1.3 | 0.03 | 0.13 | 0.14 | 0.26 | 1.26 | 0.1 |
| Butyl benzoate | 1376 1 | 1371 | 0.05 | 0.06 | 0.04 | 0.02 | |||||
| α-Copaene | 1374 2 | 1376 | 2.24 | 0.23 | 2.9 | 0.39 | 0.24 | 0.09 | 1.32 | 0.58 | 0.36 |
| Geranyl acetate | 1379 2 | 1382 | 0.05 | ||||||||
| β-Bourbonene | 1387 2 | 1384 | 0.07 | ||||||||
| (Z)-Cinnamyl acetate | 1388 2 | 1387 | 0.53 | ||||||||
| β-Cubebene | 1387 2 | 1390 | 0.06 | 0.05 | 0.02 | ||||||
| β-Elemene | 1389 2 | 1392 | 0.14 | 0.02 | 0.14 | 0.05 | |||||
| Methyl eugenol | 1403 2 | 1403 | 0.18 | 0.06 | 0.13 | ||||||
| cis-α-Bergamotene | 1411 2 | 1414 | 0.05 | ||||||||
| (E)-Caryophyllene | 1417 2 | 1420 | 2.75 | 1.53 | 0.57 | 1.55 | 1.02 | 2.68 | 2.72 | 1.45 | 1.96 |
| 2-Methylbutyl benzoate | 1438 2 | 1436 | 0.01 | 0.03 | 0.05 | 0.03 | |||||
| Aromadendrene | 1439 2 | 1438 | 0.05 | 0.02 | 0.04 | 0.11 | 0.01 | 0.03 | |||
| (E)-Cinnamyl acetate | 1443 2 | 1452 | 32.1 | 4.43 | 14.94 | 0.33 | 0.14 | 0.82 | 1.2 | 26.15 | 0.36 |
| α-Humulene | 1452 2 | 1454 | 0.7 | 0.32 | 0.09 | 0.28 | 0.21 | 0.52 | 0.68 | 0.33 | 0.38 |
| 9-epi-(E)-Caryophyllene | 1464 2 | 1460 | 0.05 | 0.03 | 0.04 | ||||||
| Cadina-1(6),4-diene | 1475 2 | 1474 | 0.05 | 0.02 | 0.02 | ||||||
| γ-Muurolene | 1478 2 | 1477 | 0.03 | 0.05 | 0.04 | 0.02 | 0.09 | 0.02 | 0.05 | ||
| Germacrene D | 1484 2 | 1481 | 0.08 | 0.12 | 0.07 | 0.83 | 0.07 | ||||
| 2-Phenylethyl 2-methylbutanoate | 1486 2 | 1485 | 0.11 | 0.02 | |||||||
| (E)-Muurola-4(14),5-diene | 1493 2 | 1492 | 0.05 | 0.03 | |||||||
| β-Selinene | 1489 2 | 1494 | 0.09 | ||||||||
| Viridiflorene | 1496 2 | 1494 | 0.09 | 0.14 | |||||||
| Bicyclogermacrene | 1500 2 | 1497 | 1.4 | 0.73 | 0.14 | 0.76 | 3.25 | 1.35 | |||
| α-Muurolene | 1500 2 | 1500 | 0.03 | 0.02 | 0.06 | 0.02 | 0.03 | 0.02 | 0.02 | ||
| β-Bisabolene | 1505 2 | 1508 | 0.04 | ||||||||
| γ-Cadinene | 1513 2 | 1514 | 0.08 | 0.15 | 0.04 | 0.02 | 0.02 | 0.06 | 0.07 | ||
| Cubebol | 1514 2 | 1515 | 0.04 | ||||||||
| δ-Cadinene | 1522 2 | 1523 | 0.33 | 0.29 | 0.53 | 0.04 | 0.13 | 0.1 | 0.25 | 0.19 | 0.23 |
| Eugenol acetate | 1521 2 | 1526 | 3.78 | 0.26 | 1.48 | 0.19 | |||||
| (E)-o-Methoxycinnamaldehyde | 1527 2 | 1527 | 0.03 | 0.03 | |||||||
| (E)-Cadina-1,4-diene | 1533 2 | 1532 | 0.03 | ||||||||
| α-Cadinene | 1537 2 | 1537 | 0.02 | ||||||||
| α-Calacorene | 1544 2 | 1542 | 0.03 | ||||||||
| Caryolan-8-ol | 1571 2 | 1569 | 0.02 | ||||||||
| Spathulenol | 1577 2 | 1577 | 0.81 | 0.15 | 3.95 | 0.05 | 0.06 | 0.19 | 0.64 | 0.8 | 0.07 |
| Caryophyllene oxide | 1582 2 | 1582 | 0.61 | 0.31 | 7.54 | 0.17 | 0.56 | 0.41 | 0.67 | 0.52 | 0.53 |
| β-Copaen-4α-ol | 1590 2 | 1586 | 0.34 | ||||||||
| Viridiflorol | 1592 2 | 1591 | 0.03 | 0.02 | 0.13 | 0.03 | |||||
| Cubeban-11-ol | 1595 2 | 1593 | 0.03 | ||||||||
| Rosifoliol | 1600 2 | 1601 | 0.08 | ||||||||
| Humulene epoxide II | 1608 2 | 1608 | 0.1 | 0.83 | 0.06 | 0.03 | 0.06 | 0.06 | 0.05 | ||
| 1-epi-Cubenol | 1627 2 | 1627 | 0.06 | 0.17 | 0.04 | 0.02 | |||||
| Caryophylla-4(12),8(13)-dien-5-α-ol | 1639 2 | 1631 | 0.05 | ||||||||
| Caryophylla-4(12),8(13)-dien-5-β-ol | 1639 2 | 1635 | 0.02 | 0.43 | 0.02 | 0.08 | |||||
| α-Muurolol (=Torreyol) | 1644 2 | 1640 | 0.14 | ||||||||
| epi-α-Murrolol (=τ-muurolol) | 1640 2 | 1640 | 0.08 | ||||||||
| epi-α-Cadinol (=τ-cadinol) | 1638 2 | 1640 | 0.2 | 0.04 | 0.06 | ||||||
| Cubenol | 1645 2 | 1642 | 0.06 | 0.07 | |||||||
| α-Cadinol | 1652 2 | 1654 | 0.03 | 0.12 | 0.03 | 0.04 | 0.03 | 0.07 | 0.07 | ||
| 14-Hydroxy-9-epi-(E)-caryophyllene | 1668 2 | 1670 | 0.45 | ||||||||
| Mustakone | 1676 2 | 1676 | 0.07 | ||||||||
| Khusinol | 1675 2 | 1684 | 0.12 | ||||||||
| Amorpha-4,9-dien-2-ol | 1704 2 | 1698 | 0.04 | ||||||||
| Benzyl benzoate | 1759 2 | 1769 | 15.83 | 76.51 | 44.11 | 0.29 | 1.22 | 22.96 | 68.16 | 47.68 | 0.28 |
| Phytone | 1841 1 | 1843 | 0.02 | 0.02 | |||||||
| 2-Phenylethyl benzoate | 1856 1 | 1852 | 0.37 | 2.43 | 0.03 | 0.23 | 0.11 | ||||
| Benzyl salicylate | 1864 2 | 1866 | 0.15 | 0.03 | 0.02 | ||||||
| Phytol | 2110 1 | 2110 | 0.06 | ||||||||
| Monoterpene Hydrocarbons | 8.08 | 3.81 | 4.77 | 0.63 | 0.75 | 6.01 | 7.24 | 1.83 | 0.92 | ||
| Oxygenated Monoterpenes | 2.79 | 4.24 | 1.45 | 0.04 | 0.67 | 3.56 | 4.47 | 1.06 | 2.51 | ||
| Sesquiterpene Hydrocarbons | 8.15 | 3,2 | 4.74 | 2.42 | 1.89 | 4.38 | 8.98 | 4.97 | 3.31 | ||
| Oxygenated Sesquiterpenes | 1.76 | 0.72 | 14.07 | 0.22 | 0.71 | 0.73 | 1.79 | 1.64 | 0.8 | ||
| Phenylpropanoids | 56.4 | 9.25 | 21.94 | 96.14 | 93.89 | 60.01 | 6.57 | 40.11 | 91.37 | ||
| Benzenoids | 21.66 | 78.71 | 50.46 | 0.53 | 1.75 | 25.05 | 70.32 | 49.13 | 0.67 | ||
| Others | 0.12 | 0.2 | 0.18 | 0.03 | 0.13 | 0.1 | 0.03 | ||||
| Total Identifield | 98.96 | 99.93 | 97.63 | 99.98 | 99.84 | 99.77 | 99.5 | 98.84 | 99.61 | ||
2.2. DNA Authentication and Genetic Variability of Cinnamomum verum Samples
2.3. Molecular and Chemical Methods
3. Materials and Methods
3.1. Plant Material
3.2. Essential Oil Extraction
3.3. GC-MS and GC(FID) Analysis
3.4. Multivariate Statistical Analysis of Chemical Composition
3.5. DNA Isolation, PCR Amplification, and Sequencing
3.6. Sequence Identity and Distance Genetics Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bandusekara, B.S.; Pushpakumara, D.K.N.G.; Bandaranayake, P.C.G.; Wijesinghe, K.G.G.; Jayasinghe, G.G. Field Level Identification of Cinnamomum Species in Sri Lanka Using a Morphological Index. Trop. Agric. Res. 2020, 31, 43–53. [Google Scholar] [CrossRef]
- WFO. World Flora Online. 2022. Available online: http://www.worldfloraonline.org (accessed on 15 April 2022).
- Rani, A.; Pande, C.; Tewari, G.; Patni, K. A review on aroma profile of cinnamomum species in north and north east India. World J. Pharm. Res. 2017, 6, 200–221. [Google Scholar] [CrossRef] [Green Version]
- Lopes, J.D.S.; de Lima, A.B.S.; da Cruz Cangussu, R.R.; da Silva, M.V.; Ferrão, S.P.B.; Santos, L.S. Application of spectroscopic techniques and chemometric methods to differentiate between true cinnamon and false cinnamon. Food Chem. 2022, 368, 130746. [Google Scholar] [CrossRef]
- Suriyagoda, L.; Mohotti, A.J.; Vidanarachchi, J.K.; Kodithuwakku, S.P.; Chathurika, M.; Bandaranayake, P.C.; Hetherington, A.M.; Beneragama, C.K. “Ceylon cinnamon”: Much more than just a spice. Plants People Planet 2021, 3, 319–336. [Google Scholar] [CrossRef]
- Abeysekera, W.P.K.M.; Premakumara, G.A.S.; Ratnasooriya, W.D. In vitro antioxidant properties of leaf and bark extracts of ceylon cinnamon (Cinnamomum zeylanicum Blume). Trop. Agric. Res. 2013, 24, 128–138. [Google Scholar]
- Ministry of Development Strategies and International Trade (MDSIT) and Sri Lanka Export Development Board (SLEDB). Government of Sri Lanka National Export Strategy of Sri Lanka—Spices and Concentrates Strategy. 13–14; Government of Sri Lanka: Sri Lanka, 2018. Available online: www.sputnix.es (accessed on 14 February 2022).
- Singh, N.; Rao, A.S.; Nandal, A.; Kumar, S.; Yadav, S.S.; Ganaie, S.A.; Narasimhan, B. Phytochemical and pharmacological review of Cinnamomum verum J. Presl-a versatile spice used in food and nutrition. Food Chem. 2021, 338, 127773. [Google Scholar] [CrossRef]
- Sadeghi, S.; Davoodvandi, A.; Pourhanifeh, M.H.; Sharifi, N.; ArefNezhad, R.; Sahebnasagh, R.; Moghadam, S.A.; Sahebkar, A.; Mirzaei, H. Anti-cancer effects of cinnamon: Insights into its apoptosis effects. Eur. J. Med. Chem. 2019, 178, 131–140. [Google Scholar] [CrossRef]
- Zare, R.; Nadjarzadeh, A.; Zarshenas, M.M.; Shams, M.; Heydari, M. Efficacy of cinnamon in patients with type II diabetes mellitus: A randomized controlled clinical trial. Clin. Nutr. 2019, 38, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Ranasinghe, P.; Galappaththy, P.; Constantine, G.R.; Jayawardena, R.; Weeratunga, H.D.; Premakumara, S.; Katulanda, P. Cinnamomum zeylanicum (Ceylon cinnamon) as a potential pharmaceutical agent for type-2 diabetes mellitus: Study protocol for a randomized controlled trial. Trials 2017, 18, 446. [Google Scholar] [CrossRef] [PubMed]
- Gulcin, I.; Kaya, R.; Goren, A.C.; Akincioglu, H.; Topal, M.; Bingol, Z.; Alwasel, S. Anticholinergic, antidiabetic and antioxidant activities of cinnamon (Cinnamomum verum) bark extracts: Polyphenol contents analysis by LC-MS/MS. Int. J. Food Prop. 2019, 22, 1511–1526. [Google Scholar] [CrossRef] [Green Version]
- Schink, A.; Naumoska, K.; Kitanovski, Z.; Kampf, C.J.; Fröhlich-Nowoisky, J.; Thines, E.; Lucas, K. Anti-inflammatory effects of cinnamon extract and identification of active compounds influencing the TLR2 and TLR4 signaling pathways. Food Funct. 2018, 9, 5950–5964. [Google Scholar] [CrossRef] [PubMed]
- Semenya, S.S.; Potgieter, M.J.; Erasmus, L.J.C. Ethnobotanical survey of medicinal plants used by Bapedi traditional healers to manage HIV/AIDS in the Limpopo Province, South Africa. J. Med. Plant Res. 2013, 7, 434–441. [Google Scholar] [CrossRef]
- Padalia, H.; Rathod, T.; Moteriya, P.; Chanda, S. Antimicrobial efficacy of Cinnamonum verum essential oil alone and in combination with antibiotics and other essential oils. Int. J. Curr. Microbiol. Appl. 2017, 6, 3377–3395. [Google Scholar] [CrossRef]
- Al-Zereini, W.A.; Al-Trawneh, I.N.; Al-Qudah, M.A.; TumAllah, H.M.; Al-Rawashdeh, H.A.; Abudayeh, Z.H. Essential oils from Elettaria cardamomum (L.) Maton grains and Cinnamomum verum J. Presl barks: Chemical examination and bioactivity studies. J. Pharm. Pharmacogn. Res. 2022, 10, 173–185. [Google Scholar] [CrossRef]
- Jirovetz, L.; Buchbauer, G.; Ruzicka, J.; Shafi, M.P.; Rosamma, M.K. Analysis of Cinnamomum zeylanicum Blume leaf oil from South India. J. Essent. Oil Res. 2001, 13, 442–443. [Google Scholar] [CrossRef]
- Farias, A.P.P.; Monteiro, O.D.S.; da Silva, J.K.R.; Figueiredo, P.L.B.; Rodrigues, A.A.C.; Monteiro, I.N.; Maia, J.G.S. Chemical composition and biological activities of two chemotype-oils from Cinnamomum verum J. Presl growing in North Brazil. J. Food Sci. Technol. 2020, 57, 3176–3183. [Google Scholar] [CrossRef]
- Binitha Raj, R.V.; Rajesh, K.S.; Mahadevan, S.; Rosamma, M.P.; Meena, C.V. Detection of adulteration in commercial samples of Cinnamomum verum JS Presl from Kerala. Eur. J. Biomed. 2018, 5, 297–304. [Google Scholar]
- Geethakumary, M.P.; Pandurangan, A.G.; Santhoshkumar, E.S. Cinnamomum litseaefolium (Lauraceae)–A new distributional record for India. Rheedea 2012, 22, 127–130. [Google Scholar]
- Ananthakrishnan, R.; Santhoshkumar, E.S.; Rameshkumar, K.B. Comparative chemical profiles of essential oil constituents of eight wild Cinnamomum species from the Western Ghats of India. Nat. Prod. Commun. 2018, 13, 1934578X1801300525. [Google Scholar] [CrossRef] [Green Version]
- Xavier, J.K.A.; Maia, L.; Figueiredo, P.L.B.; Folador, A.; Ramos, A.R.; Andrade, E.H.; Maia, J.G.S.; Setzer, W.N.; da Silva, J.K.R. Essential Oil Composition and DNA Barcode and Identification of Aniba species (Lauraceae) Growing in the Amazon Region. Molecules 2021, 26, 1914. [Google Scholar] [CrossRef]
- da Trindade, R.C.S.; Xavier, J.K.A.M.; Setzer, W.N.; Maia, J.G.S.; da Silva, J.K.R. Chemical Diversity and Therapeutic Effects of Essential Oils of Aniba Species from the Amazon: A Review. Plants 2021, 10, 1854. [Google Scholar] [CrossRef]
- Santhosh Kumar, J.U.; Krishna, V.; Seethapathy, G.S.; Ganesan, R.; Ravikanth, G.; Shaanker, R.U. Assessment of adulteration in raw herbal trade of important medicinal plants of India using DNA barcoding. 3 Biotech 2018, 8, 135. [Google Scholar] [CrossRef]
- Li, Y.; Geng, L.; Liu, Y.; Chen, M.; Mu, Q.; Zhang, X.; Ren, G.; Liu, C. Identification of three Daphne species by DNA barcoding and HPLC fingerprint analysis. PLoS ONE 2018, 13, e0201711. [Google Scholar] [CrossRef]
- Yu, J.; Wu, X.; Liu, C.; Newmaster, S.; Ragupathy, S.; Kress, W.J. Progress in the use of DNA barcodes in the identification and classification of medicinal plants. Ecotoxicol. Environ. Saf. 2021, 208, 111691. [Google Scholar] [CrossRef]
- Swetha, V.P.; Parvathy, V.A.; Sheeja, T.E.; Sasikumar, B. DNA barcoding for discriminating the economically important Cinnamomum verum from its adulterants. Food Biotechnol. 2014, 28, 183–194. [Google Scholar] [CrossRef]
- Fahlén, A.; Welander, M.; Wennersten, R. Effects of light–temperature regimes on plant growth and essential oil yield of selected aromatic plants. J. Sci. Food. Agric. 1997, 73, 111–119. [Google Scholar] [CrossRef]
- Malsawmtluangi, L.; Nautiyal, B.P.; Hazarika, T.; Chauhan, R.S.; Tava, A. Essential oil composition of bark and leaves of Cinammoum verum Bertch. & Presl from Mizoram, North East India. J. Essent. Oil Res. 2016, 28, 551–556. [Google Scholar] [CrossRef]
- Koketsu, M.; Gonçalves, S.L.; Godoy, R.L.D.O.; Lopes, D.; Morsbach, N. Óleos essenciais de cascas e folhas de canela (Cinnamomum verum Presl) cultivada no Paraná. Food Sci. Technol. 1997, 17, 281–285. [Google Scholar] [CrossRef]
- Chinh, H.V.; Luong, N.X.; Thin, D.B.; Dai, D.N.; Hoi, T.M.; Ogunwande, I.A. Essential oils leaf of Cinnamomum glaucescens and Cinnamomum verum from Vietnam. J. Essent. Oil Res. 2017, 8, 2712. [Google Scholar] [CrossRef] [Green Version]
- Ariyarathne, H.B.M.A.; Weerasooriya, S.N.; Senarath, W.T.P.S.K. Comparison of morphological and chemical characteristics of two selected accessions and six wild species of genus Cinnamomum Schaeff. Sri Lankan J. Biol. 2018, 3, 11–23. [Google Scholar] [CrossRef] [Green Version]
- Wijeweera, A.A.; Hewage, J.W.; Jayasinghe, G.G.; Waduthanthrige, S.H.; Hettiarachchi, S.R.; Wijesinghe, K.G. Maturity dependence of quality, quantity and chemical constituents of bark and leaf oil of Ceylon Cinnamon (Cinnamomum zeylanicum Blume). Ruhuna J. Sci. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Li, Y.; Kong, D.; Lin, X.; Xie, Z.; Bai, M.; Huang, S.; Nian, H.; Wu, H. Quality evaluation for essential oil of Cinnamomum verum leaves at different growth stages based on GC–MS, FTIR and microscopy. Food Anal. Methods 2016, 9, 202–212. [Google Scholar] [CrossRef]
- Monteiro, I.N.; dos Santos Monteiro, O.; Costa-Junior, L.M.; da Silva Lima, A.; de Aguiar Andrade, E.H.; Maia, J.G.S.; Mouchrek Filho, V.E. Chemical composition and acaricide activity of an essential oil from a rare chemotype of Cinnamomum verum Presl on Rhipicephalus microplus (Acari: Ixodidae). Vet. Parasitol. 2017, 238, 54–57. [Google Scholar] [CrossRef]
- Liyanage, N.N.; Ranawake, A.L.; Bandaranayake, P.C.G. Cross-pollination effects on morphological, molecular, and biochemical diversity of a selected cinnamon (Cinnamomum zeylanicum Blume) seedling population. J. Crop Improv. 2021, 35, 21–37. [Google Scholar] [CrossRef]
- Chakraborty, A.; Sankaran, V.; Ramar, M.; Chellappan, D.R. Chemical analysis of leaf essential oil of Cinnamomum verum from Palni hills, Tamil Nadu. J. Chem. Pharm. 2015, 8, 476–479. [Google Scholar]
- Jantan, I.B.; Karim Moharam, B.A.; Santhanam, J.; Jamal, J.A. Correlation between chemical composition and antifungal activity of the essential oils of eight cinnamomum. Species. Pharm. Biol. 2008, 46, 406–412. [Google Scholar] [CrossRef] [Green Version]
- Dongmo, P.M.J.; Tatsadjieu, L.N.; Tchoumbougnang, F.; Sameza, M.L.; Dongmo, B.N.; Zollo, P.H.A.; Menut, C. Chemical composition, antiradical and antifungal activities of essential oil of the leaves of Cinnamomum zeylanicum Blume from Cameroon. Nat. Prod. Commun. 2007, 2, 1934578X0700201219. [Google Scholar] [CrossRef] [Green Version]
- de Castro, C.C.; da Silva, A.R.C.; Franco, C.D.J.P.; Siqueira, G.M.; Cascaes, M.M.; do Nascimento, L.D.; de Aguiar Andrade, E.H. Caracterização química do óleo essencial das folhas, galhos e frutos de Cinnamomum verum J. Presl (Lauraceae). Braz. J. Dev. 2020, 6, 41320–41333. [Google Scholar] [CrossRef]
- Lima, M.D.P.; Zoghbi, M.D.G.B.; Andrade, E.H.A.; Silva, T.M.D.; Fernandes, C.S. Constituintes voláteis das folhas e dos galhos de Cinnamomum zeylanicum Blume (Lauraceae). Acta Amaz 2005, 35, 363–366. [Google Scholar] [CrossRef] [Green Version]
- ISO 3524:2003(E); Oil of Cinnamon Leaf, Sri Lanka Type (Cinnamomum zeylanicum Blume). ISO (International Organization for Standardization): Geneva, Switzerland, 2003.
- Boniface, Y.; Philippe, S.; de Lima, H.R.; Pierre, N.J.; Alain, A.G.; Fatiou, T.; Dominique, S. Chemical composition and antimicrobial activities of Cinnamomum zeylanicum Blume dry leaves essential oil against food-borne pathogens and adulterated microorganisms. Int. Res. J. Biol. Sci. 2012, 1, 18–25. [Google Scholar]
- Jayaprakasha, G.K.; Jagan Mohan Rao, L.; Sakariah, K.K. Chemical composition of the flower oil of Cinnamomum zeylanicum Blume. J. Agric. Food Chem. 2000, 48, 4294–4295. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.S.; Liu, J.Y.; Hsui, Y.R.; Chang, S.T. Chemical polymorphism and antifungal activity of essential oils from leaves of different provenances of indigenous cinnamon (Cinnamomum osmophloeum). Bioresour. Technol. 2006, 97, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Csikós, E.; Csekő, K.; Ashraf, A.R.; Kemény, Á.; Kereskai, L.; Kocsis, B.; Böszörményi, A.; Helyes, Z.; Horváth, G. Effects of Thymus vulgaris L., Cinnamomum verum J. Presl and Cymbopogon nardus (L.) rendle essential oils in the endotoxin-induced acute airway inflammation mouse model. Molecules 2020, 25, 3553. [Google Scholar] [CrossRef]
- Andrade, M.A.; Cardoso, M.D.G.; Batista, L.R.; Mallet, A.C.T.; Machado, S.M.F. Óleos essenciais de Cymbopogon nardus, Cinnamomum zeylanicum e Zingiber officinale: Composição, atividades antioxidante e antibacteriana. Rev. Cienc. Agron. 2012, 43, 399–408. [Google Scholar] [CrossRef]
- Nath, S.C.; Pathak, M.G.; Baruah, A. Benzyl benzoate, the major component of the leaf and stem bark oil of Cinnamomum zeylanicum Blume. J. Essent. Oil Res. 1996, 8, 327–328. [Google Scholar] [CrossRef]
- Raina, V.K.; Srivastava, S.K.; Aggarwal, K.K.; Ramesh, S.; Kumar, S. Essential oil composition of Cinnamomum zeylanicum Blume leaves from Little Andaman, India. Flavour Fragr. J. 2001, 16, 374–376. [Google Scholar] [CrossRef]
- Adams, R.P. Identification of Essential Oil Components by Gas Chromatography/Mass Spectrometry; Allured Publishing Corporation: Carol Stream, IL, USA, 2007. [Google Scholar]
- Mondello, L. FFNSC 2: Flavors and Fragrances of Natural and Synthetic Compounds, Mass Spectral Database; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2011. [Google Scholar]
- Alberts, P.S.F.; Meyer, J.J.M. Integrating chemotaxonomic-based metabolomics data with DNA barcoding for plant identification: A case study on south-east African Erythroxylaceae species. S. Afr. J. Bot. 2022, 146, 174–186. [Google Scholar] [CrossRef]
- El-Atroush, H. DNA Barcoding of Two Medicinal Plants Using Molecular Markers. Egypt. Acad. J. Biolog. Sci. (C. Physiol. Mol. Biol.) 2020, 12, 83–92. [Google Scholar]
- Kress, W.J.; Erickson, D.L. A two-locus global DNA barcode for land plants: The coding RBCL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef] [Green Version]
- CBOL Plant Working Group 1; Hollingsworth, P.M.; Forrest, L.L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; van der Bank, M.; Chase, M.W.; Cowan, R.S.; Little, D.P.; et al. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef] [Green Version]
- Kiran, K.R.; Swathy, P.S.; Paul, B.; Prasada, K.S.; Rao, M.R.; Joshi, M.B.; Rai, P.S.R.; Satyamoorthy, K.; Muthusamy, A. Untargeted metabolomics and DNA barcoding for discrimination of Phyllanthus species. J. Ethnopharmacol. 2021, 273, 113928. [Google Scholar] [CrossRef] [PubMed]
- Seethapathy, G.S.; Ganesh, D.; Santhosh Kumar, J.U.; Senthilkumar, U.; Newmaster, S.G.; Ragupathy, S.; Shaanker, R.U.; Ravikanth, G. Assessing product adulteration in natural health products for laxative yielding plants, Cassia, Senna, and Chamaecrista, in Southern India using DNA barcoding. Int. J. Leg. Med. 2015, 129, 693–700. [Google Scholar] [CrossRef]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and using a plant DNA barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J.; Erickson, D.L.; Jones, F.A.; Swenson, N.G.; Perez, R.; Sanjur, O.; Bermingham, E. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc. Natl. Acad. Sci. USA 2009, 106, 18621–18626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trofimov, D.; Rudolph, B.; Rohwer, J.G. Phylogenetic study of the genus Nectandra (Lauraceae), and reinstatement of Damburneya. Taxon 2016, 65, 980–996. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekara, C.B.; Naranpanawa, D.N.U.; Bandusekara, B.S.; Pushpakumara, D.K.N.G.; Wijesundera, D.S.A.; Bandaranayake, P.C. Universal barcoding regions, rbcL, matK and trnH-psbA do not discriminate Cinnamomum species in Sri Lanka. PLoS ONE 2021, 16, e0245592. [Google Scholar] [CrossRef]
- Yang, B.C.; Lee, M.S.; Sun, F.C.; Chao, H.H.; Chang, W.T.; Lin, M.K.; Chen, H.J.; Lee, M.S. Rapid identification of the indigenous medicinal crop Cinnamomum osmophloeum from various adulterant Cinnamomum species by DNA polymorphism analysis. Pharmacogn. Mag. 2020, 16, 64. [Google Scholar]
- Sánchez, M.; González-Burgos, E.; Divakar, P.K.; Gómez-Serranillos, M.P. DNA-based authentication and metabolomics analysis of medicinal plants samples by DNA barcoding and ultra-high-performance liquid chromatography/triple quadrupole mass spectrometry (UHPLC-MS). Plants 2020, 9, 1601. [Google Scholar] [CrossRef]
- Feitosa-Alcantara, R.B.; Arrigoni-Blank, M.D.F.; Blank, A.F.; Nogueira, P.C.D.L.; Sampaio, T.S.; Nizio, D.A.D.C.; Almeida-Pereira, C.S. Chemical diversity of essential oils from Hyptis pectinata (L.) Poit. Biosci. J. 2018, 34, 875–887. [Google Scholar] [CrossRef] [Green Version]
- Wink, M. Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry 2003, 64, 3–19. [Google Scholar] [CrossRef]
- Philippe, F.; Dubrulle, N.; Marteaux, B.; Bonnet, B.; Choisy, P.; Berthon, J.Y.; Garnier, L.; Leconte, N.; Milesi, S.; Morvan, P.Y.; et al. Combining DNA Barcoding and Chemical fingerprints to authenticate Lavender raw material. Int. J. Cosmet. Sci. 2022, 44, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Kumar, A.; Nagireddy, A.; Mani, D.N.; Shukla, A.K.; Tiwari, R.; Sundaresan, V. DNA barcoding: An efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnol. J. 2016, 14, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Cristians, S.; Bye, R.; Nieto-Sotelo, J. Molecular markers associated with chemical analysis: A powerful tool for quality control assessment of copalchi medicinal plant complex. Front. Pharmacol. 2018, 9, 666. [Google Scholar] [CrossRef] [Green Version]
- van den Dool, H.; Kratz, P.D. A generalization of the retention index system including linear temperature programmed gas-liquid partition chromatography. J. Chromatogr. 1963, A11, 463–471. [Google Scholar] [CrossRef]
- Lias, S.G.; Mikaia, A.I.; Sparkman, O.D.; Stein, S.E.; Zaikin, G. The NIST/EPA/NIH Mass Spectral Database: Simultaneous Control of Quality and Quantity. In Proceedings of the 45th ASMS Conference on Mass Spectrometry and Allied Topics, Palm Springs, CA, USA, 1–5 June 1997; Available online: https://www.nist.gov/publications/nistepanih-mass-spectral-database-simultaneous-control-quality-and-quantity (accessed on 26 February 2022).
- Liu, Z.F.; Ci, X.Q.; Li, L.; Li, H.W.; Conran, J.G.; Li, J. DNA barcoding evaluation and implications for phylogenetic relationships in Lauraceae from China. PLoS ONE 2017, 12, e0175788. [Google Scholar] [CrossRef] [Green Version]
- Sang, T.; Crawford, D.J.; Stuessy, T.F. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 1997, 84, 1120–1136. [Google Scholar] [CrossRef] [Green Version]
- Tate, J.A.; Simpson, B.B. Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Syst. Bot. 2003, 28, 723–737. [Google Scholar] [CrossRef]
- Ford, C.S.; Ayres, K.L.; Toomey, N.; Haider, N.; Van Alphen Stahl, J.; Kelly, L.J.; Wikstrom, N.; Hollingsworth, P.M.; Duff, R.J.; Hoot, S.B.; et al. Selection of candidate coding DNA barcoding regions for use on land plants. Bot. J. Linn. 2009, 159, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kimura, M.A. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]


| DNA Markers | Aligned Length (bp) | Nucleotide Diversity (π) | Polymorphic Sites | Parsimony-Informative Sites |
|---|---|---|---|---|
| rbcL | 543 | 0.000123 | 3 | 0 |
| matK | 738 | 0.00000 | 0 | 0 |
| psbA-trnH | 418 | 0.01449 | 20 | 6 |
| rbcL+matK+psbA-trnH | 1699 | 0.00700 | 23 | 6 |
| Sample Code | Type | Collection Site/Company | Voucher |
|---|---|---|---|
| Cve1 | Cultivated | Maranhãozinho (MA) | 243613 |
| Cve2 | Cultivated | Curuçá (PA) | 243614 |
| Cve3 | Cultivated | Belém (PA) | 243615 |
| Cve4 | Cultivated | Benevides (PA) | 243616 |
| Cve5 | Cultivated | Belém (PA) | 243617 |
| Cve6-c | commercialized | Ver-o-peso market | Not cataloged |
| Cve7-c | commercialized | Tempero e Cia | Not cataloged |
| CVe8-c | commercialized | Ver-o-peso market | Not cataloged |
| Cve9-c | commercialized | Pau de verônica | Not cataloged |
| Region | Primers | Sequence (5′–3′) | Amplification Protocol |
|---|---|---|---|
| matK 1 | matk 2.1 | CCTATCCATCTGGAAATCTTAG | 95 °C 7min; 95 °C 1min, 53 °C 1 min, 72 °C 1 min, 35 cycles; 72 °C 7 min |
| matk 5 | GTTCTAGCACAAGAAAGTCG | ||
| psbA 2 | psbA3_f F | GTTATGCATGAACGTAATGCT | 95 °C 7 min; 95 °C 1min, 56 °C 1 min, 72 °C 1 min, 35 cycles; 72 °C 7 min |
| trnH 3 | trnHf_05 R | CGCGCATGGTGGATTCACAATCC | |
| rbcL 4 | rbcL1 | ATGTCACCACAAACAGAGACTAAAGC | 95 °C 7min; 95 °C 1min, 53 °C 1 min, 72 °C 1 min, 35 cycles; 72 °C 7 min |
| rbcLa | GTAAAATCAAGTCCACCRCG |
| Sample Code | matK | psbA-trnH | rbcL |
|---|---|---|---|
| Cve1 | OM981169 | OM981164 | OM981159 |
| Cve2 | OM981170 | OM981165 | OM981160 |
| Cve3 | OM981171 | OM981166 | OM981161 |
| Cve4 | OM981172 | OM981167 | OM981162 |
| Cve5 | OM981173 | OM981168 | OM981163 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xavier, J.K.A.M.; Baia, T.G.C.; Alegria, O.V.C.; Figueiredo, P.L.B.; Carneiro, A.R.; Moreira, E.C.d.O.; Maia, J.G.S.; Setzer, W.N.; da Silva, J.K.R. Essential Oil Chemotypes and Genetic Variability of Cinnamomum verum Leaf Samples Commercialized and Cultivated in the Amazon. Molecules 2022, 27, 7337. https://doi.org/10.3390/molecules27217337
Xavier JKAM, Baia TGC, Alegria OVC, Figueiredo PLB, Carneiro AR, Moreira ECdO, Maia JGS, Setzer WN, da Silva JKR. Essential Oil Chemotypes and Genetic Variability of Cinnamomum verum Leaf Samples Commercialized and Cultivated in the Amazon. Molecules. 2022; 27(21):7337. https://doi.org/10.3390/molecules27217337
Chicago/Turabian StyleXavier, Júlia Karla A. M., Talissa Gabriele C. Baia, Oscar Victor C. Alegria, Pablo Luis B. Figueiredo, Adriana R. Carneiro, Edith Cibelle de O. Moreira, José Guilherme S. Maia, William N. Setzer, and Joyce Kelly R. da Silva. 2022. "Essential Oil Chemotypes and Genetic Variability of Cinnamomum verum Leaf Samples Commercialized and Cultivated in the Amazon" Molecules 27, no. 21: 7337. https://doi.org/10.3390/molecules27217337
APA StyleXavier, J. K. A. M., Baia, T. G. C., Alegria, O. V. C., Figueiredo, P. L. B., Carneiro, A. R., Moreira, E. C. d. O., Maia, J. G. S., Setzer, W. N., & da Silva, J. K. R. (2022). Essential Oil Chemotypes and Genetic Variability of Cinnamomum verum Leaf Samples Commercialized and Cultivated in the Amazon. Molecules, 27(21), 7337. https://doi.org/10.3390/molecules27217337