Ru(II)(ƞ6-p-cymene) Conjugates Loaded onto Graphene Oxide: An Effective pH-Responsive Anticancer Drug Delivery System
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis and Characterization
2.2. In Vitro DNA Interaction Studies
2.3. DNA Cleavage Studies by Electrophoretic Gel Assay
2.4. Computational Studies
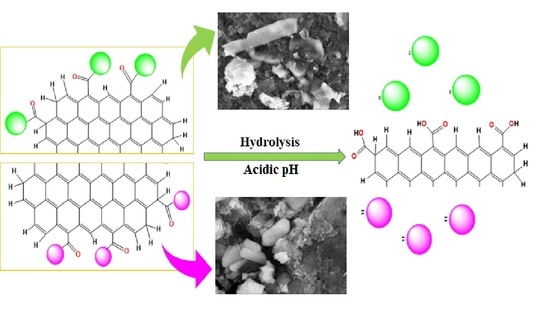
2.5. Drug Loading and Release Profile
2.6. Cytotoxicity
3. Materials and Method
3.1. Materials
3.2. Instrumentation
3.3. Synthesis
3.4. Cytotoxicity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
References
- Tabassum, S.; Pettinari, C. Chemical and Biotechnological Developments in Organotin Cancer Chemotherapy. J. Organomet. Chem. 2006, 691, 1761–1766. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA. Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Zaki, M.; Afzal, M.; Ahmad, M.; Tabassum, S. Synthesis and Crystal Structure Elucidation of New Copper(II)-Based Chemotherapeutic Agent Coupled with 1,2-DACH and Orthovaniline: Validated by in Vitro DNA/HSA Binding Profile and PBR322 Cleavage Pathway. J. Photochem. Photobiol. B Biol. 2016, 161, 318–327. [Google Scholar] [CrossRef]
- Parveen, S.; Arjmand, F.; Tabassum, S. Development and Future Prospects of Selective Organometallic Compounds as Anticancer Drug Candidates Exhibiting Novel Modes of Action. Eur. J. Med. Chem. 2019, 175, 269–286. [Google Scholar] [CrossRef]
- Ortega, E.; Vigueras, G.; Ballester, F.J.; Ruiz, J. Targeting Translation: A Promising Strategy for Anticancer Metallodrugs. Coord. Chem. Rev. 2021, 446, 214129. [Google Scholar] [CrossRef]
- Ghosh, S. Cisplatin: The First Metal Based Anticancer Drug. Bioorg. Chem. 2019, 88, 102925. [Google Scholar] [CrossRef]
- Welsh, A.; Rylands, L.I.; Arion, V.B.; Prince, S.; Smith, G.S. Synthesis and Antiproliferative Activity of Benzimidazole-Based, Trinuclear Neutral Cyclometallated and Cationic, N^N-Chelated Ruthenium(Ii) Complexes. Dalt. Trans. 2020, 49, 1143–1156. [Google Scholar] [CrossRef]
- Montani, M.; Pazmay, G.V.B.; Hysi, A.; Lupidi, G.; Pettinari, R.; Gambini, V.; Tilio, M.; Marchetti, F.; Pettinari, C.; Ferraro, S.; et al. The Water Soluble Ruthenium(II) Organometallic Compound [Ru(p-Cymene)(Bis(3,5 Dimethylpyrazol-1-Yl)Methane)Cl]Cl Suppresses Triple Negative Breast Cancer Growth by Inhibiting Tumor Infiltration of Regulatory T Cells. Pharmacol. Res. 2016, 107, 282–290. [Google Scholar] [CrossRef]
- Fleige, E.; Quadir, M.A.; Haag, R. Stimuli-Responsive Polymeric Nanocarriers for the Controlled Transport of Active Compounds: Concepts and Applications. Adv. Drug Deliv. Rev. 2012, 64, 866–884. [Google Scholar] [CrossRef]
- Oliveira, A.M.L.; Machado, M.; Silva, G.A.; Bitoque, D.B.; Ferreira, J.T.; Pinto, L.A.; Ferreira, Q. Graphene Oxide Thin Films with Drug Delivery Function. Nanomaterials 2022, 12, 1149. [Google Scholar] [CrossRef]
- Cao, M.; Zhao, W.; Wang, L.; Li, R.; Gong, H.; Zhang, Y.; Xu, H.; Lu, J.R. Graphene Oxide-Assisted Accumulation and Layer-by-Layer Assembly of Antibacterial Peptide for Sustained Release Applications. ACS Appl. Mater. Interfaces 2018, 10, 24937–24946. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Wu, Z.; Li, N.; Pu, Y.; Wang, B.; Zhang, T.; Tao, J. Advanced Review of Graphene-Based Nanomaterials in Drug Delivery Systems: Synthesis, Modification, Toxicity and Application. Mater. Sci. Eng. C 2017, 77, 1363–1375. [Google Scholar] [CrossRef]
- McCallion, C.; Burthem, J.; Rees-Unwin, K.; Golovanov, A.; Pluen, A. Graphene in Therapeutics Delivery: Problems, Solutions and Future Opportunities. Eur. J. Pharm. Biopharm. 2016, 104, 235–250. [Google Scholar] [CrossRef]
- Muschi, M.; Serre, C. Progress and Challenges of Graphene Oxide/Metal-Organic Composites. Coord. Chem. Rev. 2019, 387, 262–272. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Zheng, Y.; Tan, C.P.; Sun, J.H.; Zhang, W.; Ji, L.N.; Mao, Z.W. Graphene Oxide Decorated with Ru(II)-Polyethylene Glycol Complex for Lysosome-Targeted Imaging and Photodynamic/Photothermal Therapy. ACS Appl. Mater. Interfaces 2017, 9, 6761–6771. [Google Scholar] [CrossRef]
- Wang, Y.; Qiu, M.; Won, M.; Jung, E.; Fan, T.; Xie, N.; Chi, S.G.; Zhang, H.; Kim, J.S. Emerging 2D Material-Based Nanocarrier for Cancer Therapy beyond Graphene. Coord. Chem. Rev. 2019, 400, 213041. [Google Scholar] [CrossRef]
- Wei, L.; Li, G.; Lu, T.; Wei, Y.; Nong, Z.; Wei, M.; Pan, X.; Qin, Q.; Meng, F.; Li, X. Functionalized Graphene Oxide as Drug Delivery Systems for Platinum Anticancer Drugs. J. Pharm. Sci. 2021, 110, 3631–3638. [Google Scholar] [CrossRef]
- Liu, Z.; Robinson, J.T.; Sun, X.; Dai, H. PEGylated Nanographene Oxide for Delivery of Water-Insoluble Cancer Drugs. J. Am. Chem. Soc. 2008, 130, 10876–10877. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.K.; Melchart, M.; Habtemariam, A.; Sadler, P.J. Organometallic Chemistry, Biology and Medicine: Ruthenium Arene Anticancer Complexes. Chem. Commun. 2005, 4764–4776. [Google Scholar] [CrossRef]
- Alessio, E. Thirty Years of the Drug Candidate NAMI-A and the Myths in the Field of Ruthenium Anticancer Compounds: A Personal Perspective. Eur. J. Inorg. Chem. 2017, 2017, 1549–1560. [Google Scholar] [CrossRef]
- Clarke, M.J.; Zhu, F.; Frasca, D.R. Non-Platinum Chemotherapeutic Metallopharmaceuticals. Chem. Rev. 1999, 99, 2511–2533. [Google Scholar] [CrossRef]
- Liang, X.; Jiang, J.; Xue, X.; Huang, L.; Ding, X.; Nong, D.; Chen, H.; Pan, L.; Ma, Z. Synthesis, Characterization, Photoluminescence, Anti-Tumor Activity, DFT Calculations and Molecular Docking with Proteins of Zinc(Ii) Halogen Substituted Terpyridine Compounds. Dalt. Trans. 2019, 48, 10488–10504. [Google Scholar] [CrossRef]
- Akhter, S.; Rehman, A.; Abidi, S.M.A.; Arjmand, F.; Tabassum, S. Synthesis, Structural Insights, and Biological Screening of DNA Targeted Ru(Ii)(H6-p-Cymene) Complexes Containing Bioactive Amino-Benzothiazole Ligand Scaffolds. N. J. Chem. 2022, 46, 11462–11473. [Google Scholar] [CrossRef]
- Huang, J.; Wang, D.; Yue, Z.; Li, X.; Chu, D.; Yang, P. Ruthenium Dye N749 Covalently Functionalized Reduced Graphene Oxide: A Novel Photocatalyst for Visible Light H2 Evolution. J. Phys. Chem. C 2015, 119, 27892–27899. [Google Scholar] [CrossRef]
- Lai, L.; Chen, L.; Zhan, D.; Sun, L.; Liu, J.; Lim, S.H.; Poh, C.K.; Shen, Z.; Lin, J. One-Step Synthesis of NH2-Graphene from in Situ Graphene-Oxide Reduction and Its Improved Electrochemical Properties. Carbon 2011, 49, 3250–3257. [Google Scholar] [CrossRef]
- Kavitha, T.; Kang, I.K.; Park, S.Y. Poly(Acrylic Acid)-Grafted Graphene Oxide as an Intracellular Protein Carrier. Langmuir 2014, 30, 402–409. [Google Scholar] [CrossRef]
- Farivar, F.; Lay Yap, P.; Karunagaran, R.U.; Losic, D. Thermogravimetric Analysis (TGA) of Graphene Materials: Effect of Particle Size of Graphene, Graphene Oxide and Graphite on Thermal Parameters. C 2021, 7, 41. [Google Scholar] [CrossRef]
- Su, H.; Wu, S.; Li, Z.; Huo, Q.; Guan, J.; Kan, Q. Co(II), Fe(III) or VO(II) Schiff Base Metal Complexes Immobilized on Graphene Oxide for Styrene Epoxidation. Appl. Organomet. Chem. 2015, 29, 462–467. [Google Scholar] [CrossRef]
- Holder, C.F.; Schaak, R.E. Tutorial on Powder X-Ray Diffraction for Characterizing Nanoscale Materials. ACS Nano 2019, 13, 7359–7365. [Google Scholar] [CrossRef] [Green Version]
- Dongil, A.B.; Bachiller-Baeza, B.; Guerrero-Ruiz, A.; Rodríguez-Ramos, I. Graphite Oxide as Support for the Immobilization of Ru-BINAP: Application in the Enantioselective Hydrogenation of Methylacetoacetate. Catal. Commun. 2012, 26, 149–154. [Google Scholar] [CrossRef]
- Dorniani, D.; Kura, A.U.; Hussein, M.Z.B.; Fakurazi, S.; Shaari, A.H.; Ahmad, Z. Controlled-Release Formulation of Perindopril Erbumine Loaded PEG-Coated Magnetite Nanoparticles for Biomedical Applications. J. Mater. Sci. 2014, 49, 8487–8497. [Google Scholar] [CrossRef] [Green Version]
- Akhter, S.; Usman, M.; Arjmand, F.; Tabassum, S. Synthesis, Structural Characterization, in Vitro Comparative DNA/RNA Binding, and Computational Studies of Half-Sandwich Ru (II)(ƞ6-p-Cymene) Aminoquinoline Complex. Polyhedron 2022, 213, 115618. [Google Scholar] [CrossRef]
- Sankarganesh, M.; Raja, J.D.; Revathi, N.; Solomon, R.V.; Kumar, R.S. Gold(III) Complex from Pyrimidine and Morpholine Analogue Schiff Base Ligand: Synthesis, Characterization, DFT, TDDFT, Catalytic, Anticancer, Molecular Modeling with DNA and BSA and DNA Binding Studies. J. Mol. Liq. 2019, 294, 111655. [Google Scholar] [CrossRef]
- Massoni, M.; Clavijo, J.C.T.; Colina-Vegas, L.; Villarreal, W.; Dias, J.S.M.; da Silva, G.A.F.; Ionta, M.; Soares, M.; Ellena, J.; Dorigueto, A.C.; et al. Propyl Gallate Metal Complexes: Circular Dichroism, BSA-Binding, Antioxidant and Cytotoxic Activity. Polyhedron 2017, 129, 214–221. [Google Scholar] [CrossRef]
- Zehra, S.; Roisnel, T.; Arjmand, F. Enantiomeric Amino Acid Schiff Base Copper(II) Complexes as a New Class of RNA-Targeted Metallo-Intercalators: Single X-Ray Crystal Structural Details, Comparative in Vitro DNA/RNA Binding Profile, Cleavage, and Cytotoxicity. ACS Omega 2019, 4, 7691–7705. [Google Scholar] [CrossRef]
- Khursheed, S.; Siddique, H.R.; Tabassum, S.; Arjmand, F. Water Soluble Transition Metal [Ni(Ii), Cu(Ii) and Zn(Ii)] Complexes of N-Phthaloylglycinate Bis(1,2-Diaminocyclohexane). DNA Binding, PBR322 Cleavage and Cytotoxicity. Dalt. Trans. 2022, 51, 11713–11729. [Google Scholar] [CrossRef]
- Parveen, S.; Tabassum, S.; Arjmand, F. Synthesis of Chiral: R / S -Pseudopeptide-Based Cu(II) & Zn(II) Complexes for Use in Targeted Delivery for Antitumor Therapy: Enantiomeric Discrimination with CT-DNA and PBR322 DNA Hydrolytic Cleavage Mechanism. RSC Adv. 2017, 7, 6587–6597. [Google Scholar] [CrossRef] [Green Version]
- Parveen, S.; Usman, M.; Tabassum, S.; Arjmand, F. Synthesis and Characterization of Co(II) and Fe(II) Peptide Conjugates as Hydrolytic Cleaving Agents and Their Preferential Enantiomeric Disposition for CT-DNA: Structural Investigation of l-Enantiomers by DFT and Molecular Docking Studies. RSC Adv. 2015, 5, 72121–72131. [Google Scholar] [CrossRef]
- Arjmand, F.; Afsan, Z.; Roisnel, T. Design, Synthesis and Characterization of Novel Chromone Based-Copper(Ii) Antitumor Agents with N,N-Donor Ligands: Comparative DNA/RNA Binding Profile and Cytotoxicity. RSC Adv. 2018, 8, 37375–37390. [Google Scholar] [CrossRef] [Green Version]
- Gopalakrishnan, D.; Srinath, S.; Baskar, B.; Bhuvanesh, N.S.P.; Ganeshpandian, M. Biological and Catalytic Evaluation of Ru(II)-p-Cymene Complexes of Schiff Base Ligands: Impact of Ligand Appended Moiety on Photo-Induced DNA and Protein Cleavage, Cytotoxicity and C-H Activation. Appl. Organomet. Chem. 2019, 33, e4756. [Google Scholar] [CrossRef]
- Sathyaraj, G.; Kiruthika, M.; Weyhermüller, T.; Unni Nair, B. Oxidative Cleavage of DNA by Ruthenium(II) Complexes Containing a Ferrocene/Non-Ferrocene Conjugated Imidazole Phenol Ligand. Organometallics 2012, 31, 6980–6987. [Google Scholar] [CrossRef]
- Arjmand, F.; Khursheed, S.; Roisnel, T.; Siddique, H.R. Copper (II)-Based Halogen-Substituted Chromone Antitumor Drug Entities: Studying Biomolecular Interactions with Ct-DNA Mediated by Sigma Hole Formation and Cytotoxicity Activity. Bioorg. Chem. 2020, 104, 104327. [Google Scholar] [CrossRef]
- Hussien, M.A. Novel Oxidovanadium ( IV ) Complexes as Colon Anticancer Agents. Molecules 2022, 27, 649. [Google Scholar]
- Khursheed, S.; Rafiq Wani, M.; Shadab, G.G.H.A.; Tabassum, S.; Arjmand, F. Synthesis, Structure Elucidation by Multi-Spectroscopic Techniques and Single-Crystal X-Ray Diffraction of Promising Fluoro/Bromo-Substituted-Chromone(Bpy)Copper(II) Anticancer Drug Entities. Inorg. Chim. Acta 2022, 538, 120967. [Google Scholar] [CrossRef]
- Manea, Y.K.; Khan, A.M.T.; Qashqoosh, M.T.A.; Wani, A.A.; Shahadat, M. Ciprofloxacin-Supported Chitosan/Polyphosphate Nanocomposite to Bind Bovine Serum Albumin: Its Application in Drug Delivery. J. Mol. Liq. 2019, 292, 111337. [Google Scholar] [CrossRef]
- Almurshedi, A.S.; Radwan, M.; Omar, S.; Alaiya, A.A.; Badran, M.M.; Elsaghire, H.; Saleem, I.Y.; Hutcheon, G.A. A Novel PH-Sensitive Liposome to Trigger Delivery of Afatinib to Cancer Cells: Impact on Lung Cancer Therapy. J. Mol. Liq. 2018, 259, 154–166. [Google Scholar] [CrossRef]
- Wei, P.; Gangapurwala, G.; Pretzel, D.; Leiske, M.N.; Wang, L.; Hoeppener, S.; Schubert, S.; Brendel, J.C.; Schubert, U.S. Smart PH-Sensitive Nanogels for Controlled Release in an Acidic Environment. Biomacromolecules 2019, 20, 130–140. [Google Scholar] [CrossRef]
- Liang, J.; Chen, B.; Hu, J.; Huang, Q.; Zhang, D.; Wan, J.; Hu, Z.; Wang, B. PH and Thermal Dual-Responsive Graphene Oxide Nanocomplexes for Targeted Drug Delivery and Photothermal-Chemo/Photodynamic Synergetic Therapy. ACS Appl. Bio. Mater. 2019, 2, 5859–5871. [Google Scholar] [CrossRef]
- Sava, G.; Capozzi, I.; Clerici, K.; Gagliardi, G.; Alessio, E.; Mestroni, G. Pharmacological Control of Lung Metastases of Solid Tumours by a Novel Ruthenium Complex. Clin. Exp. Metastasis 1998, 16, 371–379. [Google Scholar] [CrossRef]
- Grguipka, S.-Š.; Ivanović, I.; Rakić, G.; Todorović, N.; Gligorijević, N.; Radulović, S.; Arion, V.B.; Keppler, B.K.; Tešić, Ž.L. Ruthenium(II)-Arene Complexes with Functionalized Pyridines: Synthesis, Characterization and Cytotoxic Activity. Eur. J. Med. Chem. 2010, 45, 1051–1058. [Google Scholar] [CrossRef]
- Ivanovic, I.; Grgurić-Šipka, S.; Gligorijević, N.; Radulović, S.; Roller, A.; Tešić, Ž.L.; Keppler, B.K. X-Ray Structure and Cytotoxic Activity of a Picolinate Ruthenium (II)– Arene Complex. J. Serbian Chem. Soc. 2011, 76, 53–61. [Google Scholar] [CrossRef]
- Gligorijević, N.; Aranđelović, S.; Filipović, L.; Jakovljević, K.; Janković, R.; Grgurić-Šipka, S.; Ivanović, I.; Radulović, S.; Tešić, Ž.L. Picolinate Ruthenium(II)-Arene Complex with in Vitro Antiproliferative and Antimetastatic Properties: Comparison to a Series of Ruthenium(II)-Arene Complexes with Similar Structure. J. Inorg. Biochem. 2012, 108, 53–61. [Google Scholar] [CrossRef]
- Chelopo, M.P.; Pawar, S.A.; Sokhela, M.K.; Govender, T.; Kruger, H.G.; Maguire, G.E.M. Anticancer Activity of Ruthenium(II) Arene Complexes Bearing 1,2,3,4-Tetrahydroisoquinoline Amino Alcohol Ligands. Eur. J. Med. Chem. 2013, 66, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Y.; Zhang, J.; Sun, Y.M.; Zhu, C.F.; Lu, Y.N.; Wu, J.Z.; Li, J.; Liu, H.Y.; Ye, Y. Photodynamic Antitumor Activity of Ru(Ii) Complexes of Imidazo-Phenanthroline Conjugated Hydroxybenzoic Acid as Tumor Targeting Photosensitizers. J. Mater. Chem. B 2020, 8, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Li, J.; Zhang, S.; Guo, L.; He, X.; Kong, D.; Zhang, H.; Liu, Z. Half-Sandwich Ruthenium(Ii) Complexes Containing N^N-Chelated Imino-Pyridyl Ligands That Are Selectively Toxic to Cancer Cells. Chem. Commun. 2017, 53, 12810–12813. [Google Scholar] [CrossRef]
- Li, L.; Wong, Y.S.; Chen, T.; Fan, C.; Zheng, W. Ruthenium Complexes Containing Bis-Benzimidazole Derivatives as a New Class of Apoptosis Inducers. Dalt. Trans. 2012, 41, 1138–1141. [Google Scholar] [CrossRef]
- Yousuf, S.; Arjmand, F.; Siddique, H.R.; Ali, M.S.; Al-Lohedan, H.A.; Tabassum, S. Biophysical Binding Profile with Ct-DNA and Cytotoxic Studies of a Modulated Nanoconjugate of Umbelliferone Cobalt Oxide Loaded on Graphene Oxide (GO) as Drug Carrier. J. Biomol. Struct. Dyn. 2020, 40, 4558–4569. [Google Scholar] [CrossRef]


















| Nanocomposite | Kb (M−1) | ΔG (kJ mol−1) | Ksv (M−1) |
|---|---|---|---|
| GO-NCD-1 | 0.57 × 104 (±0.0416) M−1 | −21.40 (±1.782) kJ mol−1 | 0.168 × 103 (±0.01) M−1 |
| GO-NCD-2 | 0.32 × 105 (±0.02) M−1 | −25.7 (±0.351) kJ mol−1 | 0.153 × 103 (±0.014) M−1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akhter, S.; Arjmand, F.; Pettinari, C.; Tabassum, S. Ru(II)(ƞ6-p-cymene) Conjugates Loaded onto Graphene Oxide: An Effective pH-Responsive Anticancer Drug Delivery System. Molecules 2022, 27, 7592. https://doi.org/10.3390/molecules27217592
Akhter S, Arjmand F, Pettinari C, Tabassum S. Ru(II)(ƞ6-p-cymene) Conjugates Loaded onto Graphene Oxide: An Effective pH-Responsive Anticancer Drug Delivery System. Molecules. 2022; 27(21):7592. https://doi.org/10.3390/molecules27217592
Chicago/Turabian StyleAkhter, Suffora, Farukh Arjmand, Claudio Pettinari, and Sartaj Tabassum. 2022. "Ru(II)(ƞ6-p-cymene) Conjugates Loaded onto Graphene Oxide: An Effective pH-Responsive Anticancer Drug Delivery System" Molecules 27, no. 21: 7592. https://doi.org/10.3390/molecules27217592
APA StyleAkhter, S., Arjmand, F., Pettinari, C., & Tabassum, S. (2022). Ru(II)(ƞ6-p-cymene) Conjugates Loaded onto Graphene Oxide: An Effective pH-Responsive Anticancer Drug Delivery System. Molecules, 27(21), 7592. https://doi.org/10.3390/molecules27217592