Mechanistic Characterization of the Pharmacological Profile of HS-731, a Peripherally Acting Opioid Analgesic, at the µ-, δ-, κ-Opioid and Nociceptin Receptors
Abstract
:1. Introduction
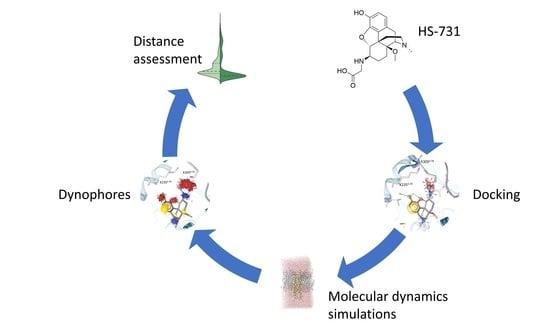
2. Results and Discussion
2.1. HS-731 Shows No Specific Binding to the NOP Receptor
2.2. Homology Modeling Is Suitable to Predict the Active State Human Nociceptin Receptor
2.3. Water Molecules Are Important for HS-731 Binding to the Opioid Receptors
2.4. Docking Reveals a Common Binding Mode for HS-731 to the Opioid Receptors
2.5. Molecular Dynamics Simulations Revealed Additional Interactions for HS-731 Binding to the Opioid Receptors
2.6. Interaction Distance Assessment Confirms Binding Hypothesis
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Cell Culture and Membrane Preparation
3.3. [3H]NOP Receptor Binding Assay
3.4. Protein Preparation and Modeling of the KOR Active Conformation
3.5. Protein-Ligand Docking
3.6. Molecular Dynamics Simulations and Analysis to Evaluate Docking Poses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Waldhoer, M.; Bartlett, S.E.; Whistler, J.L. Opioid receptors. Annu. Rev. Biochem. 2004, 73, 953–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, C. Opioid receptors. Annu. Rev. Med. 2016, 67, 433–451. [Google Scholar] [CrossRef] [PubMed]
- Corder, G.; Castro, D.C.; Bruchas, M.R.; Scherrer, G. Endogenous and exogenous opioids in pain. Annu. Rev. Neurosci. 2018, 41, 453–473. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, J.M.L.; Filizola, M. Insights From molecular dynamics simulations of a number of G-protein coupled receptor targets for the treatment of pain and opioid use disorders. Front. Mol. Neurosci. 2019, 12, 207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, A.K.; Smith, C.M.; Rahmatullah, M.; Nissapatorn, V.; Wilairatana, P.; Spetea, M.; Gueven, N.; Dietis, N. Opioid analgesia and opioid-induced adverse effects: A review. Pharmaceuticals 2021, 14, 1091. [Google Scholar] [CrossRef] [PubMed]
- Zierk, K.A. The real antidote: A critical review of U.S. and Canadian drug treatment courts and a call for public health prevention tools as a solution to the opioid epidemic. Ind. Intl Comp. L. Rev. 2019, 29, 185–217. [Google Scholar] [CrossRef]
- Sobczak, Ł.; Goryński, K. Pharmacological aspects of over-the-counter opioid drugs misuse. Molecules 2020, 25, 3905. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention; National Center for Health Statistics; Office of Communication. Drug Overdose Deaths in the U.S. Top 100,000 Annually. Available online: https://www.cdc.gov/nchs/pressroom/nchs_press_releases/2021/20211117.htm (accessed on 29 November 2021).
- Volkow, N.D.; Blanco, C. The changing opioid crisis: Development, challenges and opportunities. Mol. Psychiatry 2021, 26, 218–233. [Google Scholar] [CrossRef]
- Obeng, S.; Hiranita, T.; León, F.; McMahon, L.R.; McCurdy, C.R. Novel approaches, drug candidates, and targets in pain drug discovery. J. Med. Chem. 2021, 64, 6523–6548. [Google Scholar] [CrossRef]
- Yekkirala, A.S.; Roberson, D.P.; Bean, B.P.; Woolf, C.J. Breaking barriers to novel analgesic drug development. Nat. Rev. Drug Discov. 2017, 16, 545–564. [Google Scholar] [CrossRef] [Green Version]
- Günther, T.; Dasgupta, P.; Mann, A.; Miess, E.; Kliewer, A.; Fritzwanker, S.; Steinborn, R.; Schulz, S. Targeting multiple opioid receptors–improved analgesics with reduced side effects? Br. J. Pharmacol. 2018, 175, 2857–2868. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, C.W.; Elballa, W.M.; Vold, S.U. Bifunctional opioid receptor ligands as novel analgesics. Neuropharmacology 2019, 151, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Mores, K.L.; Cummins, B.R.; Cassell, R.J.; van Rijn, R.M. A review of the therapeutic potential of recently developed G protein-biased kappa agonists. Front. Pharmacol. 2019, 10, 407. [Google Scholar] [CrossRef] [PubMed]
- Faouzi, A.; Varga, B.R.; Majumdar, S. Biased opioid ligands. Molecules 2020, 25, 4257. [Google Scholar] [CrossRef] [PubMed]
- Bermudez, M.; Nguyen, T.N.; Omieczynski, C.; Wolber, G. Strategies for the discovery of biased GPCR ligands. Drug Discov. Today 2019, 24, 1031–1037. [Google Scholar] [CrossRef]
- Albert-Vartanian, A.; Boyd, M.; Hall, A.; Morgado, S.; Nguyen, E.; Nguyen, V.; Patel, S.; Russo, L.; Shao, A.; Raffa, R. Will peripherally restricted kappa-opioid receptor agonists (pKORA s) relieve pain with less opioid adverse effects and abuse potential? J. Clin. Pharm. Ther. 2016, 41, 371–382. [Google Scholar] [CrossRef] [Green Version]
- Martínez, V.; Abalo, R. Peripherally acting opioid analgesics and peripherally-induced analgesia. Behav. Pharmacol. 2020, 31, 136–158. [Google Scholar] [CrossRef]
- Bidlack, J.M. Detection and function of opioid receptors on cells from the immune system. Clin. Diagn. Lab. 2000, 7, 719–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holzer, P. Opioids and opioid receptors in the enteric nervous system: From a problem in opioid analgesia to a possible new prokinetic therapy in humans. Neurosci. Lett. 2004, 361, 192–195. [Google Scholar] [CrossRef]
- Machelska, H.; Celik, M.Ö. Advances in achieving opioid analgesia without side effects. Front. Pharmacol. 2018, 9, 1388. [Google Scholar] [CrossRef]
- Beck, T.C.; Hapstack, M.A.; Beck, K.R.; Dix, T.A. Therapeutic potential of kappa opioid agonists. Pharmaceuticals 2019, 12, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fürst, S.; Zádori, Z.S.; Zádor, F.; Király, K.; Balogh, M.; László, S.B.; Hutka, B.; Mohammadzadeh, A.; Calabrese, C.; Galambos, A.R. On the role of peripheral sensory and gut mu opioid receptors: Peripheral analgesia and tolerance. Molecules 2020, 25, 2473. [Google Scholar] [CrossRef] [PubMed]
- DeHaven-Hudkins, D.L.; Dolle, R.E. Peripherally restricted opioid agonists as novel analgesic agents. Curr. Pharm. Des. 2004, 10, 743–757. [Google Scholar] [CrossRef] [PubMed]
- Stokbroekx, R.A.; Vandenberk, J.; Van Heertum, A.H.; Van Laar, G.M.; Van der Aa, M.J.; Van Bever, W.F.; Janssen, P.A. Synthetic antidiarrheal agents. 2, 2-Diphenyl-4-(4’-aryl-4’-hydroxypiperidino) butyramides. J. Med. Chem. 1973, 16, 782–786. [Google Scholar] [CrossRef] [PubMed]
- Barber, A.; Bartoszyk, G.; Bender, H.; Gottschlich, R.; Greiner, H.; Harting, J.; Mauler, F.; Minck, K.O.; Murray, R.; Simon, M. A pharmacological profile of the novel, peripherally-selective κ-opioid receptor agonist, EMD 61753. Br. J. Pharmacol. 1994, 113, 1317–1327. [Google Scholar] [CrossRef]
- Spahn, V.; Del Vecchio, G.; Labuz, D.; Rodriguez-Gaztelumendi, A.; Massaly, N.; Temp, J.; Durmaz, V.; Sabri, P.; Reidelbach, M.; Machelska, H. A nontoxic pain killer designed by modeling of pathological receptor conformations. Science 2017, 355, 966–969. [Google Scholar] [CrossRef]
- Schiller, P.W. Opioid peptide-derived analgesics. In Drug Addiction, 1st ed.; Rapaka, R.S., Sadée, W., Eds.; Springer: New York, NY, USA, 2008; pp. 357–366. [Google Scholar]
- Schütz, J.; Brandt, W.; Spetea, M.; Wurst, K.; Wunder, G.; Schmidhammer, H. Synthesis of 6-amino acid Substituted derivatives of the highly potent analgesic 14-O-methyloxymorphone. Helv. Chim. Acta 2003, 86, 2142–2148. [Google Scholar] [CrossRef]
- Spetea, M.; Friedmann, T.; Riba, P.; Schütz, J.; Wunder, G.; Langer, T.; Schmidhammer, H.; Fürst, S. In vitro opioid activity profiles of 6-amino acid substituted derivatives of 14-O-methyloxymorphone. Eur. J. Pharmacol. 2004, 483, 301–308. [Google Scholar] [CrossRef]
- Fürst, S.; Riba, P.; Friedmann, T.; Tímar, J.; Al-Khrasani, M.; Obara, I.; Makuch, W.; Spetea, M.; Schütz, J.; Przewlocki, R. Peripheral versus central antinociceptive actions of 6-amino acid-substituted derivatives of 14-O-methyloxymorphone in acute and inflammatory pain in the rat. J. Pharmacol. Exp. Ther. 2005, 312, 609–618. [Google Scholar] [CrossRef] [Green Version]
- Bileviciute-Ljungar, I.; Spetea, M.; Guo, Y.; Schütz, J.; Windisch, P.; Schmidhammer, H. Peripherally mediated antinociception of the mu-opioid receptor agonist 2-(4,5alpha-epoxy-3-hydroxy-14beta-methoxy-17-methylmorphinan-6beta-yl)aminoacetic acid (HS-731) after subcutaneous and oral administration in rats with carrageenan-induced hindpaw inflammation. J. Pharmacol. Exp. Ther. 2006, 317, 220–227. [Google Scholar] [CrossRef]
- Obara, I.; Makuch, W.; Spetea, M.; Schütz, J.; Schmidhammer, H.; Przewlocki, R.; Przewlocka, B. Local peripheral antinociceptive effects of 14-O-methyloxymorphone derivatives in inflammatory and neuropathic pain in the rat. Eur. J. Pharmacol. 2007, 558, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Al-Khrasani, M.; Spetea, M.; Friedmann, T.; Riba, P.; Király, K.; Schmidhammer, H.; Furst, S. DAMGO and 6β-glycine substituted 14-O-methyloxymorphone but not morphine show peripheral, preemptive antinociception after systemic administration in a mouse visceral pain model and high intrinsic efficacy in the isolated rat vas deferens. Brain Res. Bull. 2007, 74, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Spetea, M.; Windisch, P.; Guo, Y.; Bileviciute-Ljungar, I.; Schütz, J.; Asim, M.F.; Berzetei-Gurske, I.P.; Riba, P.; Kiraly, K.; Fürst, S. Synthesis and pharmacological activities of 6-glycine substituted 14-phenylpropoxymorphinans, a novel class of opioids with high opioid receptor affinities and antinociceptive potencies. J. Med. Chem. 2011, 54, 980–988. [Google Scholar] [CrossRef]
- Baillie, L.D.; Schmidhammer, H.; Mulligan, S.J. Peripheral μ-opioid receptor mediated inhibition of calcium signaling and action potential-evoked calcium fluorescent transients in primary afferent CGRP nociceptive terminals. Neuropharmacology 2015, 93, 267–273. [Google Scholar] [CrossRef]
- Spetea, M.; Rief, S.B.; Haddou, T.B.; Fink, M.; Kristeva, E.; Mittendorfer, H.; Haas, S.; Hummer, N.; Follia, V.; Guerrieri, E. Synthesis, biological, and structural explorations of new zwitterionic derivatives of 14-O-methyloxymorphone, as potent μ/δ opioid agonists and peripherally selective antinociceptives. J. Med. Chem. 2019, 62, 641–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Botros, S.; Lipkowski, A.; Larson, D.; Stark, P.; Takemori, A.; Portoghese, P.S. Opioid agonist and antagonist activities of peripherally selective derivatives of naltrexamine and oxymorphamine. J. Med. Chem. 1989, 32, 2068–2071. [Google Scholar] [CrossRef]
- Mazak, K.; Noszal, B.; Hosztafi, S. Physicochemical and pharmacological characterization of permanently charged opioids. Curr. Med. Chem. 2017, 24, 3633–3648. [Google Scholar] [CrossRef]
- Zádor, F.; Mohammadzadeh, A.; Balogh, M.; Zádori, Z.S.; Király, K.; Barsi, S.; Galambos, A.R.; László, S.B.; Hutka, B.; Váradi, A.; et al. Comparisons of in vivo and in vitro opioid effects of newly synthesized 14-methoxycodeine-6-O-sulfate and codeine-6-O-sulfate. Molecules 2020, 25, 1370. [Google Scholar] [CrossRef] [Green Version]
- Che, T.; Majumdar, S.; Zaidi, S.A.; Ondachi, P.; McCorvy, J.D.; Wang, S.; Mosier, P.D.; Uprety, R.; Vardy, E.; Krumm, B.E.; et al. Structure of the nanobody-stabilized active state of the kappa opioid receptor. Cell 2018, 172, 55–67.e15. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Manglik, A.; Venkatakrishnan, A.J.; Laeremans, T.; Feinberg, E.N.; Sanborn, A.L.; Kato, H.E.; Livingston, K.E.; Thorsen, T.S.; Kling, R.C.; et al. Structural insights into µ-opioid receptor activation. Nature 2015, 524, 315–321. [Google Scholar] [CrossRef] [Green Version]
- Claff, T.; Yu, J.; Blais, V.; Patel, N.; Martin, C.; Wu, L.; Han, G.W.; Holleran, B.J.; van der Poorten, O.; White, K.L.; et al. Elucidating the active δ-opioid receptor crystal structure with peptide and small-molecule agonists. Sci. Adv. 2019, 5, eaax9115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballesteros, J.A.; Weinstein, H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. In Methods in Neurosciences; Academic Press: San Diego, USA; London, UK, 1995; Volume 25, pp. 366–428. [Google Scholar]
- Chavkin, C.; McLaughlin, J.P.; Celver, J.P. Regulation of opioid receptor function by chronic agonist exposure: Constitutive activity and desensitization. Mol. Pharmacol. 2001, 60, 20–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Befort, K.; Zilliox, C.; Filliol, D.; Yue, S.; Kieffer, B.L. Constitutive activation of the delta opioid receptor by mutations in transmembrane domains III and VII. J. Biol. Chem. 1999, 274, 18574–18581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Décaillot, F.M.; Befort, K.; Filliol, D.; Yue, S.; Walker, P.; Kieffer, B.L. Opioid receptor random mutagenesis reveals a mechanism for G protein-coupled receptor activation. Nat. Struct. Biol. 2003, 10, 629–636. [Google Scholar] [CrossRef]
- Váradi, A.; Marrone, G.F.; Eans, S.O.; Ganno, M.L.; Subrath, J.J.; Le Rouzic, V.; Hunkele, A.; Pasternak, G.W.; McLaughlin, J.P.; Majumdar, S. Synthesis and characterization of a dual kappa-delta opioid receptor agonist analgesic blocking cocaine reward behavior. ACS Chem. Neurosci. 2015, 6, 1813–1824. [Google Scholar] [CrossRef] [Green Version]
- Saleh, A.H.; Abdelwaly, A.; Darwish, K.M.; Eissa, A.A.H.M.; Chittiboyina, A.; Helal, M.A. Deciphering the molecular basis of the kappa opioid receptor selectivity: A molecular dynamics study. J. Mol. Graph. Model. 2021, 106, 107940. [Google Scholar] [CrossRef]
- Akuzawa, N.; Takeda, S.; Ishiguro, M. Structural modelling and mutation analysis of a nociceptin receptor and its ligand complexes. J. Biochem. 2007, 141, 907–916. [Google Scholar] [CrossRef]
- Thompson, A.A.; Liu, W.; Chun, E.; Katritch, V.; Wu, H.; Vardy, E.; Huang, X.-P.; Trapella, C.; Guerrini, R.; Calo, G.; et al. Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature 2012, 485, 395–399. [Google Scholar] [CrossRef]
- Miller, R.L.; Thompson, A.A.; Trapella, C.; Guerrini, R.; Malfacini, D.; Patel, N.; Han, G.W.; Cherezov, V.; Caló, G.; Katritch, V.; et al. The importance of ligand-receptor conformational pairs in stabilization: Spotlight on the N/OFQ G protein-coupled receptor. Structure 2015, 23, 2291–2299. [Google Scholar] [CrossRef] [Green Version]
- Mustazza, C.; Bastanzio, G. Development of nociceptin receptor (NOP) agonists and antagonists. Med. Res. Rev. 2011, 31, 605–648. [Google Scholar] [CrossRef]
- Meunier, J.-C.; Mouledous, L.; Topham, C.M. The nociceptin (ORL1) receptor: Molecular cloning and functional architecture. Peptides 2000, 21, 893–900. [Google Scholar] [CrossRef]
- Sydow, D. Dynophores: Novel Dynamic Pharmacophores Implementation of Pharmacophore Generation Based on Molecular Dynamics Trajectories and Their Graphical Representation; Freie Universität Berlin: Berlin, Germany, 2015. [Google Scholar]
- Wu, H.; Wacker, D.; Mileni, M.; Katritch, V.; Han, G.W.; Vardy, E.; Liu, W.; Thompson, A.A.; Huang, X.-P.; Carroll, F.I.; et al. Structure of the human κ-opioid receptor in complex with JDTic. Nature 2012, 485, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Fenalti, G.; Zatsepin, N.A.; Betti, C.; Giguere, P.; Han, G.W.; Ishchenko, A.; Liu, W.; Guillemyn, K.; Zhang, H.; James, D.; et al. Structural basis for bifunctional peptide recognition at human δ-opioid receptor. Nat. Struct. Mol. Biol. 2015, 22, 265–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.K.; Fitch, C.A.; García-Moreno E., B. Distance dependence and salt sensitivity of pairwise, coulombic interactions in a protein. Protein Sci. 2002, 11, 1004–1016. [Google Scholar] [CrossRef] [Green Version]
- Vardy, E.; Mosier, P.D.; Frankowski, K.J.; Wu, H.; Katritch, V.; Westkaemper, R.B.; Aubé, J.; Stevens, R.C.; Roth, B.L. Chemotype-selective modes of action of κ-opioid receptor agonists. J. Biol. Chem. 2013, 288, 34470–34483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Che, T.; English, J.; Krumm, B.E.; Kim, K.; Pardon, E.; Olsen, R.H.J.; Wang, S.; Zhang, S.; Diberto, J.F.; Sciaky, N.; et al. Nanobody-enabled monitoring of kappa opioid receptor states. Nat. Commun. 2020, 11, 1145. [Google Scholar] [CrossRef] [PubMed]
- Wacker, D.; Wang, C.; Katritch, V.; Han, G.W.; Huang, X.-P.; Vardy, E.; McCorvy, J.D.; Jiang, Y.; Chu, M.; Siu, F.Y.; et al. Structural features for functional selectivity at serotonin receptors. Science (New York, N.Y.) 2013, 340, 615–619. [Google Scholar] [CrossRef] [Green Version]
- Dumitrascuta, M.; Bermudez, M.; Trovato, O.; Neve, J.D.; Ballet, S.; Wolber, G.; Spetea, M. Antinociceptive efficacy of the µ-opioid/nociceptin peptide-based hybrid KGNOP1 in inflammatory pain without rewarding effects in mice: An experimental assessment and molecular docking. Molecules 2021, 26, 3267. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Cheng, Y.-C.; Prusoff, W.H. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. [Google Scholar] [CrossRef]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003, 10, 980. [Google Scholar] [CrossRef] [PubMed]
- Molecular Operating Environment (MOE). Molecular Operating Environment (MOE), C.C.G.U. Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7. 2021. Available online: https://www.chemcomp.com/Products.htm (accessed on 29 November 2021).
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, G.N.; Ramakrishnan, C.; Sasisekharan, V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963, 7, 95–99. [Google Scholar] [CrossRef]
- Zhu, S. Validation of the generalized force fields GAFF, CGenFF, OPLS-AA, and PRODRGFF by testing against experimental osmotic coefficient data for small drug-Like molecules. J. Chem. Inf. Model. 2019, 59, 4239–4247. [Google Scholar] [CrossRef] [PubMed]
- Bermudez, M.; Rakers, C.; Wolber, G. Structural characteristics of the allosteric binding site represent a key to subtype selective modulators of muscarinic acetylcholine receptors. Mol. Inform. 2015, 34, 526–530. [Google Scholar] [CrossRef] [PubMed]
- 3D Structure Generator CORINA Classic. Available online: https://mn-am.com/products/corina/ (accessed on 2 April 2021).
- Gasteiger, J.; Rudolph, C.; Sadowski, J. Automatic generation of 3D-atomic coordinates for organic molecules. Tetrahedron Comput. Methodol. 1990, 3, 537–547. [Google Scholar] [CrossRef]
- Labute, P. Protonate3D: Assignment of ionization states and hydrogen coordinates to macromolecular structures. Proteins 2009, 75, 187–205. [Google Scholar] [CrossRef] [Green Version]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [Green Version]
- Evers, A.; Hessler, G.; Matter, H.; Klabunde, T. Virtual screening of biogenic amine-binding G-protein coupled receptors: Comparative evaluation of protein- and ligand-based virtual screening protocols. J. Med. Chem. 2005, 48, 5448–5465. [Google Scholar] [CrossRef]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein-ligand docking using GOLD. Proteins 2003, 52, 609–623. [Google Scholar] [CrossRef]
- Vo, Q.N.; Mahinthichaichan, P.; Shen, J.; Ellis, C.R. How μ-opioid receptor recognizes fentanyl. Nat. Commun. 2021, 12, 984. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 1996, 17, 490–519. [Google Scholar] [CrossRef]
- Wolber, G.; Dornhofer, A.A.; Langer, T. Efficient overlay of small organic molecules using 3D pharmacophores. J. Comput. Aided Mol. Des. 2006, 20, 773–788. [Google Scholar] [CrossRef] [PubMed]
- Wolber, G.; Langer, T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model. 2005, 45, 160–169. [Google Scholar] [CrossRef]
- Schrödinger Release -4: Maestro, version Release -4; Schrödinger, LLC: New York, NY, USA, 2020.
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and Testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Ponder, J.W.; Case, D.A. Force fields for protein simulations. In Advances in Protein Chemistry; Academic Press: Cambridge, MA, USA, 2003; Volume 66, pp. 27–85. [Google Scholar]
- Bowers, K.J.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the Proceedings of the ACM/IEEE Conference on Supercomputing (SC06), Tampa, FL, USA, 11–17 November 2006. [Google Scholar]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Bock, A.; Bermudez, M.; Krebs, F.; Matera, C.; Chirinda, B.; Sydow, D.; Dallanoce, C.; Holzgrabe, U.; Amici, M.d.; Lohse, M.J.; et al. Ligand binding ensembles determine graded agonist efficacies at a G protein-coupled receptor. J. Biol. Chem. 2016, 291, 16375–16389. [Google Scholar] [CrossRef] [Green Version]
- van Rossum, G.; Drake, F. Python 3 Reference Manual; CreateSpace: Scotts Valley, CA, USA, 2009. [Google Scholar]
- Waskom, M. Seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]








| Receptor | Rat Opioid Receptor | Human Opioid Receptor | ||
|---|---|---|---|---|
| Binding affinity | Binding affinity | Functional activity | ||
| Ki (nM) | Ki (nM) | EC50 (nM) | % stim. | |
| MOR | 0.83 ± 0.02 a | 0.90 ± 0.14 b | 3.78 ± 0.73 b | 98 ± 9 b |
| DOR | 7.86 ± 0.64 a | 10.1 ± 2.7 b | 7.92 ± 1.63 b | 103 ± 7 b |
| KOR | 44.8 ± 0.1 a | - c | 361 ± 154 b | 82 ± 9 b |
| NOP | - c | >10,000 | - d | - d |
| InteractionType | Interaction Partners | |||
|---|---|---|---|---|
| HS-731 | MOR | DOR | KOR | |
| Cationic interaction | morphinan amine | D1493.32(100%) | D1283.32 (100%) | D1383.32 (100%) |
| Cationic interaction | secondary amine | D56N-terminus (73.7%) | Not present | E209ECL2 (12.5%) |
| Anionic interaction | Carboxylate | K2355.39 (81.3%) | K2145.39 (65.0%) | K2275.39 (63.3%) |
| K3056.58 (75.0%) | R291ECL3 (9.2%) | K200ECL2 (15.7%) | ||
| Interaction | BU72 | MP1104 | ||
|---|---|---|---|---|
| MOR | DOR | KOR | NOP | |
| PI | D1493.32 | D1283.32 | D1383.32 | D1303.32 |
| HY | M1533.36 | M1323.36 | M1423.36 | M1343.36 |
| HY | V2385.42 | V2175.42 | V2305.42 | I2195.42 |
| HY | I2986.51 | - | - | - |
| HY | V3026.55 | V2816.55 | I2946.55 | 2836.55 |
| HY | W3207.35 | - | - | - |
| HBA | - | Y1293.33 | Y1393.33 | - |
| HBA/HBD | HOH525/508 | HOH101/1301/1302 | HOH | HOH |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puls, K.; Schmidhammer, H.; Wolber, G.; Spetea, M. Mechanistic Characterization of the Pharmacological Profile of HS-731, a Peripherally Acting Opioid Analgesic, at the µ-, δ-, κ-Opioid and Nociceptin Receptors. Molecules 2022, 27, 919. https://doi.org/10.3390/molecules27030919
Puls K, Schmidhammer H, Wolber G, Spetea M. Mechanistic Characterization of the Pharmacological Profile of HS-731, a Peripherally Acting Opioid Analgesic, at the µ-, δ-, κ-Opioid and Nociceptin Receptors. Molecules. 2022; 27(3):919. https://doi.org/10.3390/molecules27030919
Chicago/Turabian StylePuls, Kristina, Helmut Schmidhammer, Gerhard Wolber, and Mariana Spetea. 2022. "Mechanistic Characterization of the Pharmacological Profile of HS-731, a Peripherally Acting Opioid Analgesic, at the µ-, δ-, κ-Opioid and Nociceptin Receptors" Molecules 27, no. 3: 919. https://doi.org/10.3390/molecules27030919
APA StylePuls, K., Schmidhammer, H., Wolber, G., & Spetea, M. (2022). Mechanistic Characterization of the Pharmacological Profile of HS-731, a Peripherally Acting Opioid Analgesic, at the µ-, δ-, κ-Opioid and Nociceptin Receptors. Molecules, 27(3), 919. https://doi.org/10.3390/molecules27030919