Phytochemical Characterizations of Maranthes polyandra (Benth.) Prance
Abstract
:1. Introduction
2. Results and Discussion
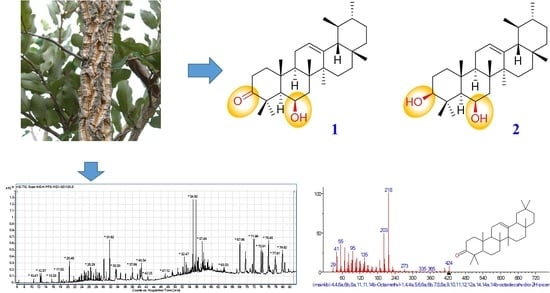
2.1. Structure Elucidation of Isolated Compounds
2.2. Phytochemical Investigation of Hexane Fraction by GC-MS
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Collection of Plant Material
3.3. Extraction and Isolation
3.4. Gas Chromatography–Mass Spectrometry (GC-MS) Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; Barreca, D.; Weckwerth, W.; Bauer, R.; Bayer, E.A.; et al. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chintamunnee, V.; Mahomoodally, M.F. Herbal medicine commonly used against non-communicable diseases in the tropical island of Mauritius. J. Herb. Med. 2012, 2, 113–125. [Google Scholar] [CrossRef]
- Gurib-Fakim, A. Medicinal plants: Traditions of yesterday and drugs of tomorrow. Mol. Aspects Med. 2006, 27, 1–93. [Google Scholar] [CrossRef] [PubMed]
- Odetoye, T.E.; Ogunniyi, D.S.; Olatunji, G.A. Refining and characterization of under-utilised seed oil of Parinari polyandra Benth for industrial utilization. Niger. J. Pure Appl. Sci. 2014, 27, 2538–2551. [Google Scholar]
- Keay, R.W.J.; Onochie, C.F.; Stanfield, D.P. Trees of Nigeria; Clarendon Press: Oxford, UK, 1989. [Google Scholar]
- Aniama, S.O.; Usman, S.S.; Ayodele, S.M. Ethnobotanical documentation of some plants among Igala people of Kogi State. Int. J. Eng. Sci. 2016, 5, 33–42. [Google Scholar]
- Allabi, A.C.; Busia, K.; Ekanmian, V.; Bakiono, F. The use of medicinal plants in self-care in the Agonlin region of Benin. J. Ethnopharmacol. 2011, 133, 234–243. [Google Scholar] [CrossRef]
- Vongtau, H.O.; Abbah, J.; Ngazal, I.E.; Kunle, O.F.; Chindo, B.A.; Otsapa, P.B.; Gamaniel, K.S. Anti-nociceptive and anti-inflammatory activities of the methanolic extract of Parinari polyandra stem bark in rats and mice. J. Ethnopharmacol. 2004, 90, 115–121. [Google Scholar] [CrossRef]
- Abolaji, A.O.; Adebayo, A.H.; Odesanmi, O.S. Effects of ethanolic fruit extract of Parinari polyandra (rosaceae) on serum lipid profile and some electrolytes in pregnant rabbits. Res. J. Med. Plant 2007, 1, 121–127. [Google Scholar] [CrossRef]
- Burkill, H.M. The Useful Plants of West Tropical Africa, 2nd ed.; Royal Botanic Gardens: Richmond, UK, 1985; ISBN 094764301X. [Google Scholar]
- Tor-Anyiin, T.A.; Anyam, J.V.; Anger, G.; Anyam, J.N. Preliminary phytochemical screening and antimicrobial activity of dried seed extracts of Maranthes polyandra. J. Chem. Soc. Niger. 2015, 40, 24–27. [Google Scholar]
- Ighodaro, O.; Omole, J.; Adejuwon, A.O.; Odunaiya, A.A. Effects of Parinari polyandra seed extract on blood glucose level and biochemical indices in Wistar Rats. Int. J. Diabetes Res. 2012, 1, 68–72. [Google Scholar] [CrossRef]
- Biosci, I.J.; Kagambega, W.; Ouattara, L.; Dibala, C.I.; Semporé, J.N.; Dicko, M.H. Determination of phenolic compounds and evaluation of the antioxidant properties of four plants from Burkina Faso. Int. J. Biosci. 2020, 17, 95–105. [Google Scholar] [CrossRef]
- Umaru, A.; Serwa, M.; Ahmed, O.M. Isolation and characterization of chemical constituent of methanol leaves extract of Maranthes polyandra and its biological potentials on some selected facultative food borne pathogens. Int. J. Green Herb. Chem. 2020, 9, 398–416. [Google Scholar] [CrossRef]
- Otun, K.O.; Olatunji, G.A.; Ajiboye, A.T.; Badeggi, U.M. Isolation and characterization of the chemical constituents of the stem bark of Parinari polyandra Benth. Int. Res. J. Pure Appl. Chem. 2014, 4, 710–717. [Google Scholar] [CrossRef]
- Odetoye, T.E.; Ogunniyi, D.S.; Olatunji, G.A. Studies on the preparation of Parinari polyandra Benth seed oil alkyd resins. J. Appl. Polym. Sci. 2013, 127, 4610–4616. [Google Scholar] [CrossRef]
- Motojesi, O.; Ogunlaja, A.S.; Amos, O. Variation in lipid composition of the seed oil Parinari polyandra Benth. Asian J. Appl. Sci. 2011, 4, 195–201. [Google Scholar] [CrossRef] [Green Version]
- Lau-Cam, C.A.; Plant, M.; Hong, B.; Herbarium, K.; Previous, N. Triterpenoids of Isertia hypoleuca leaves. Phytochemistry 1973, 12, 475–476. [Google Scholar] [CrossRef]
- Dekebo, A.; Ermias, D.; Odd, R.G.; Aasen, A.J. Triterpenes from the resin of boswellia neglecta. Bull. Chem. Soc. Ethiop. 2002, 16, 87–90. [Google Scholar] [CrossRef]
- González, A.G.; Amaro, J.; Fraga, B.M.; Luis, J.G. 3-oxo-6β-hydroxyolean-18-en-28-oic acid from orthopterygium huancuy. Phytochemistry 1983, 22, 1828–1830. [Google Scholar] [CrossRef] [Green Version]
- Ohno, N.; Mabry, T.J.; Zabelt, V.; Watson, W.H. Tetrachyrin, a new rearranged kaurenoid lactone, and diterpene acids from Tetrachyron orizabaensis and Helianthus debilis. Phytochemistry 1979, 18, 1687–1689. [Google Scholar] [CrossRef]
- Mi, K.P.; Yun-Choi, H.S.; Yong, K.K. Isolation of n-Butyl-beta-D-fructopyranoside from Gastrodia elata Blume. Nat. Prod. Sci. 2006, 12, 101–103. [Google Scholar]
- Chang, I.M.; YunChoi, H.-S.; Yamasaki, K. Revision of 13C NMR assignments of β-sitosterol and β-sitosteryl-3-O-β-D-glucopyranoside isolated from Plantago asiatica Seed. Korean J. Pharmacogn. 1981, 12, 12–14. [Google Scholar]
- Chaturvedula, V.S.P.; Prakash, I. Isolation of Stigmasterol and β-Sitosterol from the dichloromethane extract of Rubus suavissimus.pdf. Int. Curr. Pharm. J. 2012, 9, 239–242. [Google Scholar] [CrossRef] [Green Version]
- Shwe, H.H.; Thein, W.W.; Win, S.S.; Pe, N.N.; Win, T. Structural characterization of stigmasterol and β-sitosterol from the roots of Premna herbacea Roxb. Int. Eur. Ext. Enablement Sci. Eng. Manag. 2019, 7, 195–201. [Google Scholar]
- Pierre, L.L.; Moses, M.N. Isolation and Characterisation of Stigmasterol and Β -Sitosterol from Odontonema Strictum (Acanthaceae). J. Innov. Pharm. Biol. Sci. 2015, 2, 88–95. [Google Scholar]
- Wei, K.; Li, W.; Koike, K.; Pei, Y.; Chen, Y.; Nikaido, T. Complete 1H and 13C NMR assignments of two phytosterols from roots of Piper nigrum. Magn. Reson. Chem. 2004, 42, 355–359. [Google Scholar] [CrossRef]
- Kontiza, I.; Abatis, D.; Malakate, K.; Vagias, C.; Roussis, V. 3-Keto steroids from the marine organisms Dendrophyllia cornigera and Cymodocea nodosa. Steroids 2006, 71, 177–181. [Google Scholar] [CrossRef]
- Faizi, S.; Ali, M.; Saleem, R.; Irfanullah; Bibi, S. Spectral assignments and reference data complete 1H and 13C NMR assignments of stigma-5-en-3-O-β-glucoside and its acetyl derivative. Magn. Reson. Chem. 2001, 39, 399–405. [Google Scholar] [CrossRef]
- Yoo, Y.C.; Shin, B.H.; Hong, J.H.; Lee, J.; Chee, H.Y.; Song, K.S.; Lee, K.B. Isolation of fatty acids with anticancer activity from Protaetia brevitarsis larva. Arch. Pharm. Res. 2007, 30, 361–365. [Google Scholar] [CrossRef]
- Choudhary, M.I.; Siddiqui, M.; Atia-tul-Wahab; Yousuf, S.; Fatima, N.; Ahmad, M.S.; Choudhry, H. Bio-catalytic structural transformation of anti-cancer steroid, drostanolone enanthate with Cephalosporium aphidicola and Fusarium lini, and cytotoxic potential evaluation of its metabolites against certain cancer cell lines. Front. Pharmacol. 2017, 8, 900. [Google Scholar] [CrossRef] [Green Version]
- Gadallah, A.S.; Yousuf, S.; Jabeen, A.; Swilam, M.M.; Khalifa, S.A.; El-Seedi, H.R.; Choudhary, M.I. Anti-inflammatory principles from Tamarix aphylla L.: A bioassay-guided fractionation study. Molecules 2020, 25, 2994. [Google Scholar] [CrossRef] [PubMed]
- Şakul, A.A.; Okur, M.E. Beta-sitosterol and its antinociceptive mechanism action. Ankara Univ. Eczac. Fak. Derg. 2021, 45, 238–252. [Google Scholar] [CrossRef]
- Rajavel, T.; Mohankumar, R.; Archunan, G.; Ruckmani, K.; Devi, K.P. Beta sitosterol and Daucosterol (phytosterols identified in Grewia tiliaefolia) perturbs cell cycle and induces apoptotic cell death in A549 cells. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sari, D.A.; Harijono; Chang, C. Evaluation of antibacterial activities from major bioactive constituents of the whole plant of Hedyotis corymbosa. Adv. Food Sci. Sustain. Agric. Agroind. Eng. 2019, 2, 39–42. [Google Scholar] [CrossRef]
- Dionisio, K.L.; Phillips, K.; Price, P.S.; Grulke, C.M.; Williams, A.; Biryol, D.; Hong, T.; Isaacs, K.K. The Chemical and Products Database, a resource for exposure-relevant data on chemicals in consumer products. Sci. Data 2018, 5, 180125. [Google Scholar] [CrossRef] [Green Version]
- FAO/WHO Joint FAO/WHO Expert Committee on Food Additives (JECFA)—PubChem Data Source. Available online: https://www.who.int/foodsafety/areas_work/chemical-risks/jecfa/en/ (accessed on 5 February 2022).
- Roche-Molina, M.; Hardwick, B.; Sanchez-Ramos, C.; Sanz-Rosa, D.; Gewert, D.; Cruz, F.M.; Gonzalez-Guerra, A.; Andres, V.; Palma, J.A.; Ibanez, B.; et al. The pharmaceutical solvent N-methyl-2-pyrollidone (NMP) attenuates inflammation through Krüppel-like factor 2 activation to reduce atherogenesis. Sci. Rep. 2020, 10, 11636. [Google Scholar] [CrossRef]
- Chuah, X.; Okechukwu, P.; Amini, F.; Teo, S. Eicosane, pentadecane and palmitic acid: The effects in in vitro wound healing studies. Asian Pac. J. Trop. Biomed. 2018, 8, 490–499. [Google Scholar] [CrossRef]
- Zhao, C.-C.; Shao, J.-H.; Li, X.; Xu, J.; Zhang, P. Antimicrobial constituents from fruits of Ailanthus altissima SWINGLE. Arch. Pharm. Res. 2005, 28, 1147–1151. [Google Scholar] [CrossRef]
- Kaur, N.; Chaudhary, J.; Jain, A.; Kishore, L. Stigmasterol: A Comprehensive review. Int. J. Pharm. Sci. Res. 2011, 2, 2259–2265. [Google Scholar]
- de Almeida, P.D.O.; de A Boleti, A.P.; Rüdiger, A.L.; Lourenço, G.A.; da Veiga Junior, V.F.; Lima, E.S. Anti-inflammatory activity of triterpenes isolated from Protium paniculatum oil-resins. Evid. Based Complement. Alternat. Med. 2015, 2015, 293768. [Google Scholar] [CrossRef] [Green Version]
- Pornprasertpol, A.; Sereemaspun, A.; Sooklert, K.; Satirapipatkul, C.; Sukrong, S. Anticancer activity of selected Colocasia gigantia fractions. J. Med. Assoc. Thai. 2015, 98 (Suppl. 1), S98–S106. [Google Scholar] [PubMed]
- Harley, B.K.; Amponsah, I.K.; Ben, I.O.; Adongo, D.W.; Mireku-Gyimah, N.A.; Baah, M.K.; Mensah, A.Y.; Fleischer, T.C. Myrianthus libericus: Possible mechanisms of hypoglycaemic action and in silico prediction of pharmacokinetics and toxicity profile of its bioactive metabolite, friedelan-3-one. Biomed. Pharmacother. 2021, 137, 111379. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.W.; Ma, S.C.; He, Z.D.; Huang, X.S.; But, P.P.H.; Wang, H.; Chan, S.P.; Ooi, V.E.C.; Xu, H.X.; Mak, T.C.W. Molecular structures and antiviral activities of naturally occurring and modified cassane furanoditerpenoids and friedelane triterpenoids from Caesalpinia minax. Bioorg. Med. Chem. 2002, 10, 2161–2170. [Google Scholar] [CrossRef]



| No. | 1 | 2 | ||
|---|---|---|---|---|
| δH a | δC b | δH a | δC b | |
| 1a | 1.93 (o c) | 41.7 | 1.63 (o) | 40.9 |
| 1b | 1.34 (o) | 0.99 (o) | ||
| 2a | 2.74 ddd (15.5, 13.5, 6.5) | 34.5 | 1.63 (o) | 27.4 |
| 2b | 2.27 ddd (15.5, 5.0, 3.0) | 1.61 (o) | ||
| 3 | - | 216.7 | 3.14 dd (10.0, 5.0) | 79.1 |
| 4 | - | 48.7 | - | 39.6 |
| 5 | 1.22 br s | 56.4 | 0.74 br s | 55.5 |
| 6 | 4.49 br s | 69.3 | 4.55 br s | 68.7 |
| 7a | 1.81 dd (14.5, 4.0) | 40.8 | 1.79 dd (14.5, 4.0) | 40.9 |
| 7b | 1.55 (o) | 1.52 (o) | ||
| 8 | - | 39.3 | - | 39.1 |
| 9 | 1.63 dd (11.5, 5.5) | 47.3 | 1.56 dd(11.5, 6.0) | 48.0 |
| 10 | - | 36.3 | - | 36.3 |
| 11a | 2.11 ddd (18.0, 11.5, 3.0) | 23.5 | 2.05 ddd (18.0, 12.0, 3.0) | 23.3 |
| 11b | 1.99 (o) | 1.95 (o) | ||
| 12 | 5.18 dd (5.0, 3.0) | 124.5 | 5.16 dd (4.5, 3.0) | 124.8 |
| 13 | - | 139.0 | - | 138.7 |
| 14 | - | 42.8 | - | 42.7 |
| 15a | 1.89 (o) | 26.6 | 1.87 (o) | 26.6 |
| 15b | 0.97 ddd (13.0, 4.0, 2.0) | 0.96 (o) | ||
| 16a | 1.98 (o) | 28.0 | 1.98 (o) | 28.1 |
| 16b | 0.86 (o) | 0.87 (o) | ||
| 17 | - | 33.8 | - | 33.8 |
| 18 | 1.33 (o) | 59.1 | 1.32 (o) | 59.1 |
| 19 | 1.31 (o) | 39.7 | 1.32 (o) | 39.7 |
| 20 | 0.88 (o) | 39.6 | 0.87 (o) | 39.6 |
| 21a | 1.37 (o) | 31.2 | 1.37 (o) | 31.3 |
| 21b | 1.24 (o) | 1.23 (o) | ||
| 22a | 1.41 (o) | 41.5 | 1.40 (o) | 41.5 |
| 22b | 1.28 (o) | 1.28 (o) | ||
| 23 | 1.15 s | 26.0 | 1.06 s | 28.0 |
| 24 | 1.40 s | 23.9 | 1.16 s | 17.2 |
| 25 | 1.50 s | 16.7 | 1.32 s | 17.0 |
| 26 | 1.35 s | 18.9 | 1.28 s | 18.6 |
| 27 | 1.02 s | 23.3 | 1.02 s | 23.4 |
| 28 | 0.80 s | 28.7 | 0.79 s | 28.7 |
| 29 | 0.78 d (6.0) | 17.4 | 0.78 d (6.0) | 17.4 |
| 30 | 0.90 br s | 21.4 | 0.90 br s | 21.4 |
| Peak Number | RT (min) | Compound Name | Molecular Formula | Molecular Weight | Area Sum% | Compound Nature | Uses | References |
|---|---|---|---|---|---|---|---|---|
| 1. | 6.36 | 2,4-Dimethylhexane | C8H18 | 114 | 0.1 | Hydrocarbon | Flavor | [37] |
| 2. | 10.47 | 2-Heptenal | C7H12O | 112 | 0.13 | Aldehyde | Flavor | [37] |
| 3. | 12.28 | 2-Ethylhexanol | C8H18O | 130 | 0.49 | Alcohol | Dispersants, printing, dying, and paints | [38] |
| 4. | 12.57 | N-Methyl-2-pyrrolidone | C5H9NO | 99 | 0.52 | Lactam | Recover certain hydrocarbons generated in processing of petrochemicals | [39] |
| 5. | 13.00 | 2-Octenal | C8H14O | 126 | 0.06 | Aldehyde | - | |
| 6. | 14.07 | n-Nonanal | C9H18O | 142 | 0.04 | Aldehyde | - | |
| 7. | 14.49 | Methyl caprylate | C9H18O2 | 158 | 0.06 | Ester | - | |
| 8. | 15.57 | Caprylic acid | C8H16O2 | 144 | 0.44 | Fatty acid | - | |
| 9. | 17.55 | 2-Decenol | C10H18O | 154 | 0.42 | Aldehyde | Flavor | [37] |
| 10. | 17.59 | Nonanoic acid | C9H18O2 | 158 | 0.02 | Aldehyde | Flavor | [37] |
| 11. | 18.24 | 2,4-Decadienal | C9H18O2 | 158 | 0.13 | Fatty acid | Flavor | [37] |
| 12. | 18.71 | 2,4-Decanedienal | C10H16O | 152 | 0.17 | Aldehyde | Flavor | [37] |
| 13. | 19.54 | n-Decanoic acid | C10H20O2 | 172 | 0.13 | Fatty acid | - | |
| 14. | 20.46 | 3-Hydroxy-4-methoxybenzaldehyde acetate | C10H10O4 | 194 | 1.16 | Aromatic compound | Flavor | [37] |
| 15. | 22.60 | Vanillic acid methyl ester | C9H10O4 | 182 | 0.06 | Aromatic compound | Flavor | [37] |
| 16. | 23.90 | Methyl 4,7,10,13-hexadecatetraenoate | C17H26O2 | 262 | 0.35 | Fatty ester | - | |
| 17. | 25.55 | n-heptadecane | C17H36 | 240 | 0.1 | Alkane | - | |
| 18. | 27.73 | n-octadecane | C18H38 | 254 | 0.14 | Hydrocarbon | A volatile oil | [37] |
| 19. | 30.08 | 1-hexadecanol | C16H34O | 242 | 2.15 | Alcohol | - | |
| 20. | 30.66 | n-Nonadecane | C19H40 | 268 | 0.86 | Hydrocarbon | - | |
| 21. | 31.62 | n-Hexadecanoic acid methyl ester | C17H34O2 | 270 | 5.17 | Ester | - | |
| 22. | 33.5 | n-Hexadecanoic acid | C16H32O2 | 256 | 0.73 | Hydrocarbon | - | |
| 23. | 34.71 | Eicosane | C20H42 | 282 | 0.93 | Hydrocarbon | Used for the treatment of eczema | [40] |
| 24. | 37.98 | 9-Octadecen-1-ol | C18H36O | 268 | 2.36 | Alcohol | - | |
| 25. | 39.39 | 1-Heptadecanol | C17H36O | 256 | 1.24 | Alcohol | - | |
| 26. | 40.12 | Methyl linoleate | C19H34O2 | 294 | 2.07 | Fatty | Anti-inflammatory | [37] |
| 27. | 40.54 | Methyl (10E)-10-octadecenoat | C19H36O2 | 296 | 2.61 | Ester | - | |
| 28. | 40.88 | Oleic acid methyl ester | C19H36O2 | 296 | 0.39 | Ester | - | |
| 29. | 42.25 | n-Octadecanoic acid, methyl ester | C20H40O2 | 312 | 0.74 | Alcohol | Emulsifier | [37] |
| 30. | 47.12 | Eicosanol | C20H40O2 | 298 | 0.63 | Arachidyl alcohol | Emollient and thickener | [37] |
| 31. | 49.08 | Kauran-16-ol | C20H34O | 290 | 0.7 | Diterpene | - | |
| 32. | 55.31 | Methyl docosanoate | C23H46O2 | 354 | 0.62 | Ester | - | |
| 33. | 67.96 | Stigmasterol | C29H48O | 412 | 6.22 | Sterol | Anti-inflammatory, antipyretic, antiarthritic, anti-ulcer, insulin-releasing, and estrogenic effects | [34,41,42] |
| 34. | 69.53 | γ-Sitosterol | C29H50O | 414 | 2.99 | Sterol | Antidiabetic activity | [43] |
| 35. | 71.44 | β-amyrone | C30H50O | 426 | 1.42 | Triterpene | Anti-inflammatory activity | [41,44] |
| 36. | 71.88 | 4,22-Stigmastadiene-3-one | C29H46O | 410 | 8.33 | Steroid | Antimicrobial activity | [41] |
| 37. | 73.91 | Stigmast-4-en-3-one | C29H48O | 412 | 7.4 | Sterol | Hypoglycemic activity | [45] |
| 38. | 77.21 | Friedelan-3-one | C30H50O | 426 | 0.92 | Triterpene | Antimicrobial activity | [46] |
| 39. | 77.81 | 3-Methoxystigmasta-5,22-diene | C30H50O | 426 | 3.85 | Steroid | - | |
| 40. | 79.82 | β-Amyrin methyl ether | C31H52O | 440 | 7.71 | Pentacyclic triterpene | - | |
| 41. | 80.49 | 5α-Stigmastane-3,6-dione | C29H48O2 | 428 | 3.27 | Sterol | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, N.; Khan, F.-A.; Salawu, K.M.; Irshad, R.; Jabeen, A.; Zhang, C.-L.; Choudhary, M.I.; Liu, X.-M.; Wang, Y. Phytochemical Characterizations of Maranthes polyandra (Benth.) Prance. Molecules 2022, 27, 1316. https://doi.org/10.3390/molecules27041316
Ali N, Khan F-A, Salawu KM, Irshad R, Jabeen A, Zhang C-L, Choudhary MI, Liu X-M, Wang Y. Phytochemical Characterizations of Maranthes polyandra (Benth.) Prance. Molecules. 2022; 27(4):1316. https://doi.org/10.3390/molecules27041316
Chicago/Turabian StyleAli, Nida, Farooq-Ahmad Khan, Kayode Muritala Salawu, Rimsha Irshad, Almas Jabeen, Chun-Lei Zhang, Muhammad Iqbal Choudhary, Xin-Min Liu, and Yan Wang. 2022. "Phytochemical Characterizations of Maranthes polyandra (Benth.) Prance" Molecules 27, no. 4: 1316. https://doi.org/10.3390/molecules27041316
APA StyleAli, N., Khan, F. -A., Salawu, K. M., Irshad, R., Jabeen, A., Zhang, C. -L., Choudhary, M. I., Liu, X. -M., & Wang, Y. (2022). Phytochemical Characterizations of Maranthes polyandra (Benth.) Prance. Molecules, 27(4), 1316. https://doi.org/10.3390/molecules27041316