Interaction Structure and Affinity of Zwitterionic Amino Acids with Important Metal Cations (Cd2+, Cu2+, Fe3+, Hg2+, Mn2+, Ni2+ and Zn2+) in Aqueous Solution: A Theoretical Study
Abstract
:1. Introduction
2. Computational Details
3. Results and Discussion
3.1. Determining the Most Stable Structure of 20 Amino Acids
3.2. Structural Characterization of Binary Complexes
3.2.1. Acidic Amino Acids and Metal Cation
3.2.2. Alkaline Amino Acids and Metal Cation
3.2.3. Hydroxy/Sulfhydryl Amino Acids and Metal Cation Complexes
3.2.4. Aromatic Amino Acids and Metal Cation Complexes
3.2.5. Other Amino Acids and Metal Cation Complexes
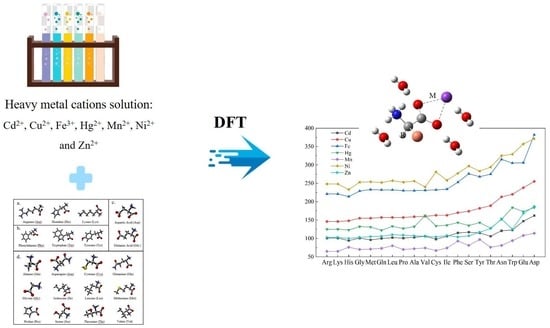
3.3. The Binding Affinity of 20 Amino Acids with Seven Metal Cations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Osuna, F.; Pavón, E.; Alba, M. Pb2+, Cd2+ and Hg2+ removal by designed functionalized swelling high-charged micas. Sci. Total Environ. 2020, 764, 142811. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liu, C.; Hsu, P.-C.; Zhao, J.; Wu, T.; Tang, J.; Liu, K.; Cui, Y. Remediation of heavy metal contaminated soil by asymmetrical alternating current electrochemistry. Nat. Commun. 2019, 10, 2440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leštan, D.; Luo, C.-L.; Li, X. The use of chelating agents in the remediation of metal-contaminated soils: A review. Environ. Pollut. 2008, 153, 3–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wuana, R.A.; Okieimen, F.E.; Imborvungu, J.A. Removal of heavy metals from a contaminated soil using organic chelating acids. Int. J. Environ. Sci. Technol. 2010, 7, 485–496. [Google Scholar] [CrossRef] [Green Version]
- Begum, Z.A.; Rahman, I.M.; Tate, Y.; Sawai, H.; Maki, T.; Hasegawa, H. Remediation of toxic metal contaminated soil by washing with biodegradable aminopolycarboxylate chelants. Chemosphere 2012, 87, 1161–1170. [Google Scholar] [CrossRef] [Green Version]
- Jelusic, M.; Lestan, D. Effect of EDTA washing of metal polluted garden soils. Part I: Toxicity hazards and impact on soil properties. Sci. Total Environ. 2014, 475, 132–141. [Google Scholar] [CrossRef]
- Gluhar, S.; Kaurin, A.; Finžgar, N.; Gerl, M.; Kastelec, D.; Lestan, D. Demonstrational gardens with EDTA-washed soil. Part I: Remediation efficiency, effect on soil properties and toxicity hazards. Sci. Total Environ. 2021, 792, 149060. [Google Scholar] [CrossRef]
- Umadevi, P.; Senthilkumar, L. Influence of metal ions (Zn2+, Cu2+, Ca2+, Mg2+ and Na+) on the water coordinated neutral and zwitterionicl-histidine dimer. RSC Adv. 2014, 4, 49040–49052. [Google Scholar] [CrossRef]
- Clark, A.A.; Yang, B.; Rodgers, M.T.; Armentrout, P.B. Experimental and Computational Study of the Group 1 Metal Cation Chelates with Lysine: Bond Dissociation Energies, Structures, and Structural Trends. J. Phys. Chem. B 2019, 123, 1983–1997. [Google Scholar] [CrossRef]
- Dolev, N.; Katz, Z.; Ludmer, Z.; Ullmann, A.; Brauner, N.; Goikhman, R. Natural amino acids as potential chelators for soil remediation. Environ. Res. 2020, 183, 109140. [Google Scholar] [CrossRef]
- Rodgers, M.T.; Armentrout, P.B.; Oomens, J.; Steill, J.D. Infrared Multiphoton Dissociation Spectroscopy of Cationized Threonine: Effects of Alkali-Metal Cation Size on Gas-Phase Conformation. J. Phys. Chem. A 2008, 112, 2258–2267. [Google Scholar] [CrossRef] [PubMed]
- Liikanen, M.; Havukainen, J.; Hupponen, M.; Horttanainen, M. Influence of different factors in the life cycle assessment of mixed municipal solid waste management systems—A comparison of case studies in Finland and China. J. Clean. Prod. 2017, 154, 389–400. [Google Scholar] [CrossRef]
- Alirezapour, F.; Khanmohammadi, A. Theoretical study on the interaction of phenylalaninal with group IA (Li+, Na+, K+) and IIA (Be2+, Mg2+, Ca2+) metal cations. J. Chin. Chem. Soc. 2021, 68, 1002–1012. [Google Scholar] [CrossRef]
- Armentrout, P.B.; Chen, Y.; Rodgers, M.T. Metal Cation Dependence of Interactions with Amino Acids: Bond Energies of Cs+ to Gly, Pro, Ser, Thr, and Cys. J. Phys. Chem. A 2012, 116, 3989–3999. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09; Revision B.01; Gaussian Inc.: Wallingford, CT, USA, 2010. [Google Scholar]
- Dennington, R.; Keith, T.; Millam, J. GAUSSVIEW; VERSION 5; Semichem Inc.: Shawnee Mission, KS, USA, 2009. [Google Scholar]
- Shankar, R.; Kolandaivel, P.; Senthilkumar, L. Interaction studies of cysteine with Li+, Na+, K+, Be2+, Mg2+, and Ca2+ metal cation complexes. J. Phys. Org. Chem. 2011, 24, 553–567. [Google Scholar] [CrossRef]
- Marenich, A.V.; Cramer, C.J.; Truhlar, D.G. Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [Google Scholar] [CrossRef]
- Grimme, S. Density functional theory with London dispersion corrections. WIREs Comput. Mol. Sci. 2011, 1, 211–228. [Google Scholar] [CrossRef]
- Gochhayat, J.K.; Dey, A.; Pathak, A.K. An ab iniio study on the micro-solvation of amino acids: On the number of water molecules necessary to stabilize the zwitter ion. Chem. Phys. Lett. 2019, 716, 93–101. [Google Scholar] [CrossRef]
- Ustunol, I.B.; Gonzalez-Pech, N.; Grassian, V.H. pH-dependent adsorption of α-amino acids, lysine, glutamic acid, serine and glycine, on TiO2 nanoparticle surfaces. J. Colloid Interface Sci. 2019, 554, 362–375. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, S.; Liu, J.; Xu, Y.; Zhou, X. Adsorption of Lysine on Na-Montmorillonite and Competition with Ca2+: A Combined XRD and ATR-FTIR Study. Langmuir 2016, 32, 4746–4754. [Google Scholar] [CrossRef]
- Kawamura, I.; Sato, H. Solid-state vibrational circular dichroism studies of L- and D-serine. Anal. Biochem. 2019, 580, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Quesada-Moreno, M.M.; Avilés-Moreno, J.R.; Márquez-García, A.A.; López-González, J.J. Deducing the molecular properties of zwitterionic, protonated, deprotonated, and double-deprotonated forms of L-cysteine from vibrational spectroscopy (IR, Raman, VCD) and quantum chemical calculations. J. Mol. Model. 2014, 20, 2229. [Google Scholar] [CrossRef] [PubMed]
- Quesada-Moreno, M.M.; Márquez-García, A.; Avilés-Moreno, J.R.; López-González, J.J. Conformational landscape of l-threonine in neutral, acid and basic solutions from vibrational circular dichroism spectroscopy and quantum chemical calculations. Tetrahedron Asymmetry 2013, 24, 1537–1547. [Google Scholar] [CrossRef]
- Stückenschneider, K.; Merz, J.; Schembecker, G. Molecular Interaction of Amino Acids with Acidic Zeolite BEA: The Effect of Water. J. Phys. Chem. C 2014, 118, 5810–5819. [Google Scholar] [CrossRef]
- Meng, L.; Hu, A.; Pang, R.; Lin, Z. Extensive Computational Study on Coordination of Transition Metal Cations and Water Molecules to Glutamic Acid. J. Phys. Chem. A 2012, 116, 7177–7188. [Google Scholar] [CrossRef]
- Khodabandeh, M.H.; Reisi, H.; Davari, M.D.; Zare, K.; Zahedi, M.; Ohanessian, G. Interaction Modes and Absolute Affinities of α-Amino Acids for Mn2+: A Comprehensive Picture. ChemPhysChem 2013, 14, 1733–1745. [Google Scholar] [CrossRef]
- Meng, L.; Lin, Z. Complexations of alkali/alkaline earth metal cations with gaseous glutamic acid. Comput. Theor. Chem. 2014, 1039, 1–10. [Google Scholar] [CrossRef]
- Xiang, F.; Bu, Y.; Ai, H.; Li, P. The Coupling Character of Ca2+ with Glutamic Acid: Implication for the Conformational Behavior and Transformation of Ca2+-ATPase in Transmembrane Ca2+ Channel. J. Phys. Chem. B 2004, 108, 17628–17638. [Google Scholar] [CrossRef]
- Heaton, A.L.; Armentrout, P.B. Experimental and Theoretical Studies of Potassium Cation Interactions with the Acidic Amino Acids and Their Amide Derivatives. J. Phys. Chem. B 2008, 112, 12056–12065. [Google Scholar] [CrossRef]
- Heaton, A.L.; Ye, S.J.; Armentrout, P.B. Experimental and Theoretical Studies of Sodium Cation Complexes of the Deamidation and Dehydration Products of Asparagine, Glutamine, Aspartic Acid, and Glutamic Acid. J. Phys. Chem. A 2008, 112, 3328–3338. [Google Scholar] [CrossRef]
- Bowman, V.N.; Heaton, A.L.; Armentrout, P.B. Metal Cation Dependence of Interactions with Amino Acids: Bond Energies of Rb+ to Gly, Ser, Thr, and Pro. J. Phys. Chem. B 2010, 114, 4107–4114. [Google Scholar] [CrossRef] [PubMed]
- Talley, J.M.; Cerda, B.A.; Ohanessian, G.; Wesdemiotis, C. Alkali Metal Ion Binding to Amino Acids Versus Their Methyl Esters: Affinity Trends and Structural Changes in the Gas Phase. Chem.—A Eur. J. 2002, 8, 1377–1388. [Google Scholar] [CrossRef]
- Hossain, M.E.; Hasan, M.M.; Halim, M.E.; Ehsan, M.Q.; Halim, M.A. Interaction between transition metals and phenylalanine: A combined experimental and computational study. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 138, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Armentrout, P.B.; Yang, B.; Rodgers, M.T. Metal Cation Dependence of Interactions with Amino Acids: Bond Energies of Rb+ and Cs+ to Met, Phe, Tyr, and Trp. J. Phys. Chem. B 2013, 117, 3771–3781. [Google Scholar] [CrossRef] [PubMed]
- Ruan, C.; Rodgers, M.T. Cation−π Interactions: Structures and Energetics of Complexation of Na+ and K+ with the Aromatic Amino Acids, Phenylalanine, Tyrosine, and Tryptophan. J. Am. Chem. Soc. 2004, 126, 14600–14610. [Google Scholar] [CrossRef]
- Bush, M.F.; Oomens, J.; Saykally, R.J.; Williams, E.R. Alkali Metal Ion Binding to Glutamine and Glutamine Derivatives Investigated by Infrared Action Spectroscopy and Theory. J. Phys. Chem. A 2008, 112, 8578–8584. [Google Scholar] [CrossRef]
- Harvey, K.B.; Porter, G.B.; Porter, G.B. Introduction to Physical Inorganic Chemistry, 3rd ed.; American Chemical Society: Washington, DC, USA, 1963; pp. 346–350. [Google Scholar]
- Jover, J.; Bosque, R.; Sales, J. A comparison of the binding affinity of the common amino acids with different metal cations. Dalton Trans. 2008, 45, 6441–6453. [Google Scholar] [CrossRef]
- Marino, T.; Toscano, M.; Russo, N.; Grand, A. Structural and Electronic Characterization of the Complexes Obtained by the Interaction between Bare and Hydrated First-Row Transition-Metal Ions (Mn2+, Fe2+, Co2+, Ni2+, Cu2+, Zn2+) and Glycine. J. Phys. Chem. B 2006, 110, 24666–24673. [Google Scholar] [CrossRef]
- Armentrout, P.B.; Yang, B.; Rodgers, M.T. Metal Cation Dependence of Interactions with Amino Acids: Bond Dissociation Energies of Rb+ and Cs+ to the Acidic Amino Acids and Their Amide Derivatives. J. Phys. Chem. B 2014, 118, 4300–4314. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Wu, M.; Li, C.; Yu, P.; Feng, S.; Li, Y.; Zhang, Q. Interaction Structure and Affinity of Zwitterionic Amino Acids with Important Metal Cations (Cd2+, Cu2+, Fe3+, Hg2+, Mn2+, Ni2+ and Zn2+) in Aqueous Solution: A Theoretical Study. Molecules 2022, 27, 2407. https://doi.org/10.3390/molecules27082407
Liu X, Wu M, Li C, Yu P, Feng S, Li Y, Zhang Q. Interaction Structure and Affinity of Zwitterionic Amino Acids with Important Metal Cations (Cd2+, Cu2+, Fe3+, Hg2+, Mn2+, Ni2+ and Zn2+) in Aqueous Solution: A Theoretical Study. Molecules. 2022; 27(8):2407. https://doi.org/10.3390/molecules27082407
Chicago/Turabian StyleLiu, Xinning, Menghan Wu, Chenchen Li, Peng Yu, Shanshan Feng, Yanwei Li, and Qingzhu Zhang. 2022. "Interaction Structure and Affinity of Zwitterionic Amino Acids with Important Metal Cations (Cd2+, Cu2+, Fe3+, Hg2+, Mn2+, Ni2+ and Zn2+) in Aqueous Solution: A Theoretical Study" Molecules 27, no. 8: 2407. https://doi.org/10.3390/molecules27082407
APA StyleLiu, X., Wu, M., Li, C., Yu, P., Feng, S., Li, Y., & Zhang, Q. (2022). Interaction Structure and Affinity of Zwitterionic Amino Acids with Important Metal Cations (Cd2+, Cu2+, Fe3+, Hg2+, Mn2+, Ni2+ and Zn2+) in Aqueous Solution: A Theoretical Study. Molecules, 27(8), 2407. https://doi.org/10.3390/molecules27082407