Synthesis and Structural Study of Amidrazone Derived Pyrrole-2,5-Dione Derivatives: Potential Anti-Inflammatory Agents
Abstract
:1. Introduction
2. Results and Discussion
2.1. The Syntheses of Compounds 2a–2f
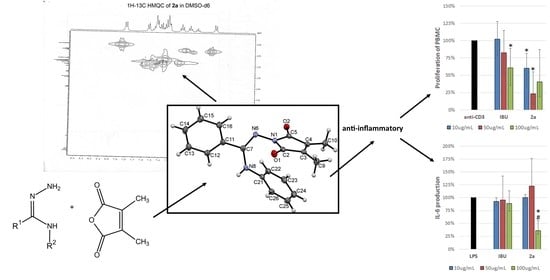
2.2. X-ray Crystallography
2.3. Toxic Activity of 2a–2f
2.4. Anti-Inflammatory Activity of 2a–2f
2.4.1. Antiproliferative Activity of 2a–2f
2.4.2. The Effects of Compounds 2a–2f on Pro-Inflammatory and Anti-Inflammatory Cytokine Production
2.5. Antibacterial Activity of 2a–2f
3. Materials and Methods
3.1. General Information
3.2. General Method of Syntheses
3.3. Crystal Structure Determination
3.4. Peripheral Blood Mononuclear Cell Preparation
3.5. In Vitro Toxic Effects on PBMCs by APC Annexin V and Propidium Iodide Staining Assay and Flow Cytometry
3.6. Anti-Inflammatory Activity
3.6.1. In Vitro Antiproliferative Effects by VPD-450 Staining Assay and Flow Cytometry
3.6.2. In Vitro Anti- and Proinflammatory Cytokine Production Effect by the Enzyme-Linked Immunosorbent Assay (ELISA)
3.7. Antibacterial Activity
3.8. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Wróbel, M.Z.; Chodkowski, A.; Herold, F.; Marciniak, M.; Dawidowski, M.; Siwek, A.; Starowicz, G.; Stachowicz, K.; Szewczyk, B.; Nowak, G.; et al. Synthesis and biological evaluation of new multi-target 3-(1H-indol-3-yl)pyrrolidine-2,5-dione derivatives with potential antidepressant effect. Eur. J. Med. Chem. 2019, 183, 111736. [Google Scholar] [CrossRef] [PubMed]
- Jan, M.S.; Ahmad, S.; Hussain, F.; Ahmad, A.; Mahmood, F.; Rashid, U.; Abid, O.-U.; Ullah, F.; Ayaz, M.; Sadiq, A. Design, synthesis, in-vitro, in-vivo and in-silico studies of pyrrolidine-2,5-dione derivatives as multitarget anti-inflammatory agents. Eur. J. Med. Chem. 2020, 186, 111863. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Alam, O.; Naim, M.J.; Shaquiquzzaman, M.; Alam, M.M.; Iqbal, M. Pyrrole: An insight into recent pharmacological advances with structure activity relationship. Eur. J. Med. Chem. 2018, 157, 527–561. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.T.; Jeon, J.Y.; Park, H.A.; Noh, Y.-S.; Lee, K.-T.; Kim, J.; Choo, D.J.; Lee, J.Y. Synthesis and PGE2 production inhibition of 1H-furan-2,5-dione and 1H-pyrrole-2,5-dione derivatives. Bioorg. Med. Chem. Lett. 2010, 20, 734–737. [Google Scholar] [CrossRef]
- Kim, K.J.; Choi, M.J.; Shin, J.-S.; Kim, M.; Choi, H.-E.; Kang, S.M.; Jin, J.H.; Lee, K.-T.; Lee, J.Y. Synthesis, biological evaluation, and docking analysis of a novel family of 1-methyl-1H-pyrrole-2,5-diones as highly potent and selective cyclooxygenase-2 (COX-2) inhibitors. Bioorg. Med. Chem. Lett. 2014, 24, 1958–1962. [Google Scholar] [CrossRef]
- Redzicka, A.; Szczukowski, L.; Kochel, A.; Wiatrak, B.; Gębczak, K.; Czyżnikowska, Z. COX-1/COX-2 inhibition activities and molecular docking study of newly designed and synthesized pyrrolo[3,4-c]pyrrole Mannich bases. Bioorg. Med. Chem. 2019, 27, 3918–3928. [Google Scholar] [CrossRef]
- Al-Zereini, W.; Yao, C.B.F.F.; Laatsch, H.; Anke, H. Aqabamycins A-G: Novel nitro maleimides from a marine Vibrio species. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. 2010, 63, 297–301. [Google Scholar] [CrossRef]
- Bycroft, B.W.; Payne, D.J. Dictionary of Antibiotics and Related Substances; CRC Press: Boca Raton, FL, USA, 2013; p. 402. [Google Scholar]
- Yuan, X.; Lu, P.; Xue, X.; Qin, H.; Fan, C.; Wang, Y.; Zhang, Q. Discovery of 2-azetidinone and 1 H -pyrrole-2,5-dione derivatives containing sulfonamide group at the side chain as potential cholesterol absorption inhibitors. Bioorg. Med. Chem. Lett. 2016, 26, 849–853. [Google Scholar] [CrossRef]
- Song, X.; Liu, C.; Chen, P.; Zhang, H.; Sun, R. Natural Product-Based Pesticide Discovery: Design, Synthesis and Bioactivity Studies of N-Amino-Maleimide Derivatives. Molecules 2018, 23, 1521. [Google Scholar] [CrossRef] [Green Version]
- Kawabe, T.; Ishigaki, M.; Sato, T.; Yamamoto, S.; Hasegawa, Y.; Canbas, Co. Ltd. Compounds with Anti-Cancer Activity. US Patent 2008275057A1, 2008. [Google Scholar]
- Chen, J.; Brooks, C.; Bergstein, I.; Stemline Therapeutics Inc. Substituted Azole Dione Compounds with Antiviral Activity. World Patent WO2021194954A1, 2021. [Google Scholar]
- SDBS—Spectral Database for Organic Compounds; National Institute of Advanced Industrial Science and Technology: Tokyo, Japan; Available online: http://sdbs.db.aist.go.jp (accessed on 25 January 2021).
- Spectra Base—Free Spectral Database; Bio-Rad Laboratories: Hercules, CA, USA; Available online: https://spectrabase.com/ (accessed on 25 January 2021).
- Cox, P.J.; Parker, S. Maleimide. Acta Crystallogr. Sect. C 1996, 52, 2578–2580. [Google Scholar] [CrossRef]
- Gill, G.B.; James, G.D.; Oates, K.V.; Pattenden, G. The synthesis of 5-ylidenepyrrol-2(5H)-ones from maleimides and from pyrrol-2-(5H)-ones. J. Chem. Soc. Perkin Trans. 1 1993, 2567–2579. [Google Scholar] [CrossRef]
- Kuehne, P.; Hesse, M. Simple synthesis of (±)-(E)-3-(4-hydroxyphenyl)-N-[4-(3-methyl-2,5-dioxo-1-pyrrolidinyl)butyl]-2-propenamide, a novel phenolic amide derivative from the bulbs of Lilium regale WILSON. Tetrahedron 1993, 49, 4575–4580. [Google Scholar] [CrossRef]
- Haddon, W.F.; Binder, R.G.; Wong, R.Y.; Harden, L.A.; Wilson, R.E.; Benson, M.; Stevens, K.L. Potent Bacterial Mutagens Produced by Chlorination of Simulated Poultry Chiller Water. J. Agric. Food Chem. 1996, 44, 256–263. [Google Scholar] [CrossRef]
- Zou, C.; Zeng, C.; Liu, Z.; Lu, M.; Sun, X.; Ye, J. γ′-Selective Functionalization of Cyclic Enones: Construction of a Chiral Quaternary Carbon Center by [4+2] Cycloaddition/Retro-Mannich Reaction with 3-Substituted Maleimides. Angew. Chem. Int. Ed. 2016, 55, 14257–14261. [Google Scholar] [CrossRef]
- Nagy, S.; Szigetvári, A.; Ilkei, V.; Krámos, B.; Béni, Z.; Szántay, C., Jr.; Hazai, L. Synthesis of aminal-type Lilium candidum alkaloids and lilaline; determination of their relative configuration by the concerted use of NMR spectroscopy and DFT conformational analysis. Tetrahedron 2021, 81, 131827. [Google Scholar] [CrossRef]
- Watson, D.J.; Dowdy, E.D.; Li, W.-S.; Wang, J.; Polniaszek, R. Electronic effects in the acid-promoted deprotection of N-2,4-dimethoxybenzyl maleimides. Tetrahedron Lett. 2001, 42, 1827–1830. [Google Scholar] [CrossRef]
- Rix, K.; Kelsall, G.H.; Hellgardt, K.; Hii, K.K.M. Chemo- and Diastereoselectivities in the Electrochemical Reduction of Maleimides. ChemSusChem 2015, 8, 665–671. [Google Scholar] [CrossRef] [Green Version]
- Schilling, W.; Zhang, Y.; Riemer, D.; Das, S. Visible-Light-Mediated Dearomatisation of Indoles and Pyrroles to Pharmaceuticals and Pesticides. Chem. Eur. J. 2020, 26, 390–395. [Google Scholar] [CrossRef] [Green Version]
- Bulatov, E.; Boyarskaya, D.; Chulkova, T.; Haukka, M. 2,3-Di-phenyl-male-imide 1-methyl-pyrrol-idin-2-one monosolvate. Acta Crystallogr. E 2014, 70, o260. [Google Scholar] [CrossRef]
- Hu, W.; Zheng, J.; Li, J.; Liu, B.; Wu, W.; Liu, H.; Jiang, H. Assembly of Polysubstituted Maleimides via Palladium-Catalyzed Cyclization Reaction of Alkynes with Isocyanides. J. Org. Chem. 2016, 81, 12451–12458. [Google Scholar] [CrossRef] [PubMed]
- Yogo, M.; Hirota, K.; Maki, Y. Synthesis of 5-iminopyrrol-2-one derivatives from 1,3-oxazines. Ring transformations via attack on the 2- or 6-position of 1,3-oxazines. J. Chem. Soc. Perkin Trans. 1 1984, 2097–2102. [Google Scholar] [CrossRef]
- Yeh, H.-C.; Wu, W.-C.; Wen, Y.-S.; Dai, D.-C.; Wang, J.-K.; Chen, C.-T. Derivative of α,β-Dicyanostilbene: Convenient Precursor for the Synthesis of Diphenylmaleimide Compounds, E−Z Isomerization, Crystal Structure, and Solid-State Fluorescence. J. Org. Chem. 2004, 69, 6455–6462. [Google Scholar] [CrossRef] [PubMed]
- Padié, C.; Zeitler, K. A novel reaction-based, chromogenic and “turn-on” fluorescent chemodosimeter for fluoride detection. New J. Chem. 2011, 35, 994–997. [Google Scholar] [CrossRef]
- Ali, A.; Siddiki, S.M.A.H.; Kon, K.; Hasegawa, J.; Shimizu, K.-I. Versatile and Sustainable Synthesis of Cyclic Imides from Dicarboxylic Acids and Amines by Nb2O5as a Base-Tolerant Heterogeneous Lewis Acid Catalyst. Chem. Eur. J. 2014, 20, 14256–14260. [Google Scholar] [CrossRef] [PubMed]
- Jafarpour, F.; Shamsianpour, M.; Issazadeh, S.; Dorrani, M.; Hazrati, H. Palladium-catalyzed direct arylation of maleimides: A simple route to bisaryl-substituted maleimides. Tetrahedron 2017, 73, 1668–1672. [Google Scholar] [CrossRef]
- Vera-Hidalgo, M.; Giovanelli, E.; Navío, C.; Pérez, E.M. Mild Covalent Functionalization of Transition Metal Dichalcogenides with Maleimides: A “Click” Reaction for 2H-MoS2 and WS2. J. Am. Chem. Soc. 2019, 141, 3767–3771. [Google Scholar] [CrossRef]
- Mendoza-Macías, C.L.; Solorio-Alvarado, C.R.; Alonso-Castro, A.J.; Alba-Betancourt, C.; Deveze-Álvarez, M.A.; Padilla-Vaca, F.; Reyes-Gualito, A. Discovery of new effective N-alkyl-3,4-diarylmaleimides-based drugs for reversing the bacterial resistance to rhodamine 6G in Bacillus subtilis. Chem. Pap. 2020, 74, 1429–1438. [Google Scholar] [CrossRef]
- Chen, P.; Cao, W.; Li, X.; Shi, D. A Unified Approach for Divergent Synthesis of Heterocycles via TMSOTf-Catalyzed Formal [3+2] Cycloaddition of Electron-Rich Alkynes. Adv. Synth. Catal. 2021, 363, 4789–4794. [Google Scholar] [CrossRef]
- Cheng, S.; Comer, D.D. An alumina-catalyzed Michael addition of mercaptans to N-anilinomaleimides and its application to the solution-phase parallel synthesis of libraries. Tetrahedron Lett. 2002, 43, 1179–1181. [Google Scholar] [CrossRef]
- Conley, N.R.; Hung, A.R.J.; Willson, C.G. A New Synthetic Route to Authentic N-Substituted Aminomaleimides. J. Org. Chem. 2005, 70, 4553–4555. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.N.; Cee, V.J.; Deak, H.L.; Du, B.; Faber, K.P.; Gunaydin, H.; Hodous, B.L.; Hollis, S.L.; Krolikowski, P.H.; Olivieri, P.R.; et al. Synthesis of 4-Substituted Chlorophthalazines, Dihydrobenzoazepinediones, 2-Pyrazolylbenzoic Acid, and 2-Pyrazolylbenzohydrazide via 3-Substituted 3-Hydroxyisoindolin-1-ones. J. Org. Chem. 2012, 77, 3887–3906. [Google Scholar] [CrossRef] [PubMed]
- Katrusiak, A.; Katrusiak, A. One-step ring condensation of hydrazine derivatives and cyclic anhydrides. J. Mol. Struct. 2015, 1085, 28–36. [Google Scholar] [CrossRef]
- Sadiq, A.; Mahnashi, M.H.; Alyami, B.A.; Alqahtani, Y.S.; Alqarni, A.O.; Rashid, U. Tailoring the substitution pattern of Pyrrolidine-2,5-dione for discovery of new structural template for dual COX/LOX inhibition. Bioorg. Chem. 2021, 112, 104969. [Google Scholar] [CrossRef] [PubMed]
- Boubekeur, K.; Grandjean, D.; Florac, C.; Robert, A. Structure of N-methoxycarbonylamino-3,4-bis(4-nitrophenyl)maleimide at 140 K. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 1991, 47, 1107–1108. [Google Scholar] [CrossRef]
- Zheng, R.; Mei, X.; Lin, Z.; Zhao, Y.; Yao, H.; Lv, W.; Ling, Q. Strong CIE activity, multi-stimuli-responsive fluorescence and data storage application of new diphenyl maleimide derivatives. J. Mater. Chem. C 2015, 3, 10242–10248. [Google Scholar] [CrossRef]
- Modzelewska, B.; Pyra, E. Synthesis of N3-substituted amidrazones. Ann. UMCS Sec. AA 1995–1996, 50/51, 111–116. [Google Scholar]
- Modzelewska-Banachiewicz, B.; Ucherek, M.; Zimecki, M.; Kutkowska, J.; Kaminska, T.; Morak-Młodawska, B.; Paprocka, R.; Szulc, M.; Lewandowski, G.; Marciniak, J.; et al. Reactions of N3-Substituted Amidrazones withcis-1,2-Cyclohexanedicarboxylic Anhydride and Biological Activities of the Products. Arch. Pharm. Chem. Life Sci. 2012, 345, 486–494. [Google Scholar] [CrossRef]
- Ziegler-Borowska, M.; Ucherek, M.; Kutkowska, J.; Mazur, L.; Modzelewska-Banachiewicz, B.; Kędziera, D.; Kaczmarek-Kędziera, A. Reaction of N3-phenylbenzamidrazone with cis-1,2-cyclohexanedicarboxylic anhydride. Tetrahedron Lett. 2010, 51, 2951–2955. [Google Scholar] [CrossRef]
- Modzelewska, B.; Banachiewicz, J.; Chodkowska, A.; Jagiełło-Wójtowicz, E.; Mazur, L. Synthesis and biological activity of new derivatives of 3-(3,4-diaryl-1,2,4-triazole-5-yl)propenoic acid. Eur. J. Med. Chem. 2004, 39, 873–877. [Google Scholar] [CrossRef]
- Paprocka, R.; Modzelewska-Banachiewicz, B.; Wiese, M.; Eljaszewicz, A.; Michalkiewicz, J. Synthesis and anti-inflammatory activity of hydrazide derivatives of 2-methylidene-1, 4-dicarboxybutanoic acid. Acta Pol. Pharm. 2012, 69, 1390–1394. [Google Scholar]
- Paprocka, R.; Wiese-Szadkowska, M.; Helmin-Basa, A.; Mazur, L.; Kutkowska, J.; Michałkiewicz, J.; Modzelewska-Banachiewicz, B.; Pazderski, L. Synthesis and evaluation of new amidrazone-derived hydrazides as a potential anti-inflammatory agents. Monatsh. Chem. 2018, 149, 1493–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paprocka, R.; Wiese, M.; Eljaszewicz, A.; Helmin-Basa, A.; Gzella, A.; Modzelewska, B.; Michalkiewicz, J. Synthesis and anti-inflammatory activity of new 1,2,4-triazole derivatives. Bioorg. Med. Chem. Lett. 2015, 25, 2664–2667. [Google Scholar] [CrossRef] [PubMed]
- Mazur, L.; Modzelewska, B.; Paprocka, R.; Zimecki, M.; Wawrzyniak, U.E.; Kutkowska, J.; Ziółkowska, G. Synthesis, crystal structure and biological activities of a novel amidrazone derivative and its copper(II) complex—A potential antitumor drug. J. Inorg. Biochem. 2012, 114, 55–64. [Google Scholar] [CrossRef]
- Mazur, L.; Sączewski, J.; Jarzembska, K.N.; Szwarc-Karabyka, K.; Paprocka, R.; Modzelewska-Banachiewicz, B. Synthesis, structural characterization and reactivity of new trisubstitutedN1-acylamidrazones: Solid state and solution studies. CrystEngComm 2018, 20, 4179–4193. [Google Scholar] [CrossRef]
- Allen, F.H. The Cambridge Structural Database: A quarter of a million crystal structures and rising. Acta Crystallogr. B 2002, 58, 380–388. [Google Scholar] [CrossRef]
- Wilson, A.J.C. International Tables for Crystallography; International Union of Crystallography: Dordrecht, Netherlands, 1992; Volume C. [Google Scholar]
- Bernstein, J.; Davis, R.E.; Shimoni, L.; Chang, N.L. Patterns in Hydrogen Bonding: Functionality and Graph Set Analysis in Crystals. Angew. Chem., Int. Ed. Engl. 1995, 32, 1555–1573. [Google Scholar] [CrossRef]
- Mooibroek, T.J.; Gamez, P.; Reedijk, J. Lone pair–π interactions: A new supramolecular bond? CrystEngComm 2008, 10, 1501–1515. [Google Scholar] [CrossRef]
- Allen, F.H.; Baalham, C.A.; Lommerse, J.P.M.; Raithby, P.R. Carbonyl-carbonyl interactions can be competitive with hydrogen bonds. Acta Crystallogr. B 1998, 54, 320–329. [Google Scholar] [CrossRef]
- Agilent Technologies. CrysAlisPRO Software System, Version 1. In 171.33.64; Oxford Diffraction Ltd.: Oxford, UK, 2010. [Google Scholar]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. A 2008, 64, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Farrugia, L.J. WinGXsuite for small-molecule single-crystal crystallography. J. Appl. Crystallogr. 1999, 32, 837–838. [Google Scholar] [CrossRef]











Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paprocka, R.; Pazderski, L.; Mazur, L.; Wiese-Szadkowska, M.; Kutkowska, J.; Nowak, M.; Helmin-Basa, A. Synthesis and Structural Study of Amidrazone Derived Pyrrole-2,5-Dione Derivatives: Potential Anti-Inflammatory Agents. Molecules 2022, 27, 2891. https://doi.org/10.3390/molecules27092891
Paprocka R, Pazderski L, Mazur L, Wiese-Szadkowska M, Kutkowska J, Nowak M, Helmin-Basa A. Synthesis and Structural Study of Amidrazone Derived Pyrrole-2,5-Dione Derivatives: Potential Anti-Inflammatory Agents. Molecules. 2022; 27(9):2891. https://doi.org/10.3390/molecules27092891
Chicago/Turabian StylePaprocka, Renata, Leszek Pazderski, Liliana Mazur, Małgorzata Wiese-Szadkowska, Jolanta Kutkowska, Michalina Nowak, and Anna Helmin-Basa. 2022. "Synthesis and Structural Study of Amidrazone Derived Pyrrole-2,5-Dione Derivatives: Potential Anti-Inflammatory Agents" Molecules 27, no. 9: 2891. https://doi.org/10.3390/molecules27092891
APA StylePaprocka, R., Pazderski, L., Mazur, L., Wiese-Szadkowska, M., Kutkowska, J., Nowak, M., & Helmin-Basa, A. (2022). Synthesis and Structural Study of Amidrazone Derived Pyrrole-2,5-Dione Derivatives: Potential Anti-Inflammatory Agents. Molecules, 27(9), 2891. https://doi.org/10.3390/molecules27092891