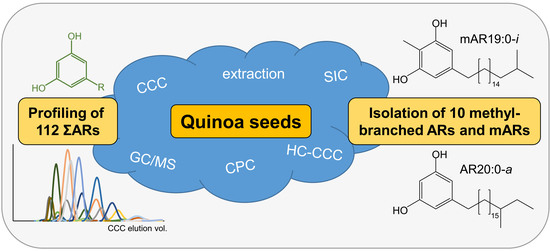
Profiling and Isolation of Ten Rare Branched-Chain Alkylresorcinols in Quinoa
Abstract
:1. Introduction
2. Results and Discussion
2.1. Enrichment of Alkylresorcinols via Centrifugal Partition Chromatography (CPC)
2.2. Profiling of ΣARs in Quinoa with Countercurrent Chromatographic Fractionation Followed by GC/MS Analysis
2.3. Isolation of Rare ∑ARs by Countercurrent Chromatography in Heart-Cut Mode (HC-CCC)—Method Description and Execution
2.3.1. Method Description
2.3.2. Description of the Fractionation with the Example of AR20:0-a with mAR19:0-i
2.3.3. HC Run A with Four Heart-Cuts (HC 1-HC 4)
2.3.4. HC run B with Two Heart-Cuts (HC 1 and HC 2)
2.4. Final Purification of (Methyl)Alkylresorcinols by Silver Ion Chromatography (SIC)
3. Materials and Methods
3.1. Quinoa Sample and Chemicals
3.2. Extraction of Alkylresorcinols (ARs) from Quinoa
3.3. Enrichment of Alkylresorcinols (ΣARs) via Centrifugal Partition Chromatography (CPC)
3.4. Isolation of Alkylresorcinols (ARs) by Countercurrent Chromatography (CCC)
3.4.1. Conventional CCC Separations (with Pool Approach Profiling)
3.4.2. Heart-Cut CCC Separations (with Pool Approach Isolation)
3.4.3. Processing of CCC Fractions
3.5. Purification of (Methyl) Alkylresorcinols ((m)ARs) by Silver Ion Chromatography
3.6. Gas Chromatography with Mass Spectrometry (GC/MS)
3.7. NMR Spectroscopy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Landberg, R.; Marklund, M.; Kamal-Eldin, A.; Åman, P. An update on alkylresorcinols—Occurrence, bioavailability, bioactivity and utility as biomarkers. J. Funct. Foods 2014, 7, 77–89. [Google Scholar] [CrossRef]
- Kozubek, A.; Tyman, J.H.P. Resorcinolic lipids, the natural non-isoprenoid phenolic amphiphiles and their biological activity. Chem. Rev. 1999, 99, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Kruk, J.; Aboul-Enein, B.; Bernstein, J.; Marchlewicz, M. Dietary alkylresorcinols and cancer prevention: A systematic review. Eur. Food Res. Technol. 2017, 243, 1693–1710. [Google Scholar] [CrossRef] [Green Version]
- Tian, S.; Sun, Y.; Chen, Z.; Zhao, R. Bioavailability and bioactivity of alkylresorcinols from different cereal products. J. Food Qual. 2020, 2020, 5781356. [Google Scholar] [CrossRef]
- Zhu, Y.; Conklin, D.R.; Chen, H.; Wang, L.; Sang, S. 5-alk(en)ylresorcinols as the major active components in wheat bran inhibit human colon cancer cell growth. Bioorg. Med. Chem. 2011, 19, 3973–3982. [Google Scholar] [CrossRef]
- Liu, L.; Winter, K.M.; Stevenson, L.; Morris, C.; Leach, D.N. Wheat bran lipophilic compounds with in vitro anticancer effects. Food Chem. 2012, 130, 156–164. [Google Scholar] [CrossRef]
- Zabolotneva, A.A.; Shatova, O.P.; Sadova, A.A.; Shestopalov, A.V.; Roumiantsev, S.A. An overview of alkylresorcinols biological properties and effects. J. Nutr. Metab. 2022, 2022, 4667607. [Google Scholar] [CrossRef]
- Kamal-Eldin, A.; Pouru, A.; Eliasson, C.; Åman, P. Alkylresorcinols as antioxidants: Hydrogen donation and peroxyl radical-scavenging effects. J. Sci. Food Agric. 2001, 81, 353–356. [Google Scholar] [CrossRef]
- El-Shabasy, R.M.; Farag, M.A. Dissecting dietary alkylresorcinols: A compile of their distribution, biosynthesis, extraction and functional properties. Crit. Rev. Biotechnol. 2023, 1–37. [Google Scholar] [CrossRef]
- Kowalska, I.; Mołdoch, J.; Pawelec, S.; Podolska, G.; von Cossel, M.; Derycke, V.; Haesaert, G.; Lana, M.A.; Lopes, M.D.S.; Riche, A.B.; et al. Environmental and cultivar variability in composition, content and biological activity of phenolic acids and alkylresorcinols of winter wheat grains from a multi-site field trial across Europe. J. Cereal Sci. 2022, 107, 103527. [Google Scholar] [CrossRef]
- Ross, A.B.; Kamal-Eldin, A.; Åman, P. Dietary alkylresorcinols: Absorption, bioactivities, and possible use as biomarkers of whole-grain wheat- and rye-rich foods. Nutr. Rev. 2004, 62, 81–95. [Google Scholar] [CrossRef]
- Marentes-Culma, R.; Orduz-Díaz, L.L.; Coy-Barrera, E. Targeted metabolite profiling-based identification of antifungal 5-n-alkylresorcinols occurring in different cereals against Fusarium oxysporum. Molecules 2019, 24, 770. [Google Scholar] [CrossRef] [Green Version]
- Luís, Â.; Domingues, F.; Duarte, A.P. Biological properties of plant-derived alkylresorcinols: Mini-review. Mini Rev. Med. Chem. 2016, 16, 851–854. [Google Scholar] [CrossRef]
- Ross, A.B.; Shepherd, M.J.; Schüpphaus, M.; Sinclair, V.; Alfaro, B.; Kamal-Eldin, A.; Åman, P. Alkylresorcinols in cereals and cereal products. J. Agric. Food Chem. 2003, 51, 4111–4118. [Google Scholar] [CrossRef]
- Pedrazzani, C.; Vanara, F.; Bhandari, D.R.; Bruni, R.; Spengler, B.; Blandino, M.; Righetti, L. 5-n-alkylresorcinol profiles in different cultivars of einkorn, emmer, spelt, common wheat, and tritordeum. J. Agric. Food Chem. 2021, 69, 14092–14102. [Google Scholar] [CrossRef]
- Zimmermann, B.F.; Patzke, H.; Schieber, A. Separation of alk(en)ylresorcinols from rye bran with saturated, monoenoic, dienoic, trienoic and hydroxylated monoenoic side chains using an octyl phase in ultra-high performance liquid chromatography and their differentiation by tandem mass spectrometrie. J. Chromatogr. A 2017, 1506, 65–72. [Google Scholar] [CrossRef]
- Hammerschick, T.; Wagner, T.; Vetter, W. Countercurrent chromatographic fractionation followed by gas chromatography/mass spectrometry identification of alkylresorcinols in rye. Anal. Bioanal. Chem. 2020, 412, 8417–8430. [Google Scholar] [CrossRef]
- Seitz, L.M. Identification of 5-(2-oxoalkyl)resorcinols and 5-(2-oxoalkenyl)resorcinols in wheat and rye grains. J. Agric. Food Chem. 1992, 40, 1541–1546. [Google Scholar] [CrossRef]
- Suzuki, Y.; Esumi, Y.; Yamaguchi, I. Structures of 5-alkylresorcinol-related analogues in rye. Phytochemistry 1999, 52, 281–289. [Google Scholar] [CrossRef]
- Ross, A.B.; Svelander, C.; Karlsson, G.; Savolainen, O.I. Identification and quantification of even and odd chained 5-n alkylresorcinols, branched chain-alkylresorcinols and methylalkylresorcinols in Quinoa (Chenopodium quinoa). Food Chem. 2017, 220, 344–351. [Google Scholar] [CrossRef]
- Hammerschick, T.; Vetter, W. Silver ion chromatography enables the separation of 2-methylalkylresorcinols from alkylresorcinols. J. Sep. Sci. 2023, unpublished. [Google Scholar]
- Racovita, R.C.; Hen-Avivi, S.; Fernandez-Moreno, J.-P.; Granell, A.; Aharoni, A.; Jetter, R. Composition of cuticular waxes coating flag leaf blades and peduncles of Triticum aestivum cv. Bethlehem. Phytochemistry 2016, 130, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Hammann, S.; Korf, A.; Bull, I.D.; Hayen, H.; Cramp, L.J.E. Lipid profiling and analytical discrimination of seven cereals using high temperature gas chromatography coupled to high resolution quadrupole time-of-flight mass spectrometry. Food Chem. 2019, 282, 27–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Hierro, J.N.; Reglero, G.; Martin, D. Chemical characterization and bioaccessibility of bioactive compounds from saponin-rich extracts and their acid-hydrolysates obtained from fenugreek and quinoa. Foods 2020, 9, 1159. [Google Scholar] [CrossRef]
- Landberg, R.; Hanhineva, K.; Tuohy, K.; Garcia-Aloy, M.; Biskup, I.; Llorach, R.; Yin, X.; Brennan, L.; Kolehmainen, M. Biomarkers of cereal food intake. Genes Nutr. 2019, 14, 28. [Google Scholar] [CrossRef]
- Ito, Y. Golden rules and pitfalls in selecting optimum conditions for high-speed counter-current chromatography. J. Chromatogr. A 2005, 1065, 145–168. [Google Scholar] [CrossRef]
- Bojczuk, M.; Żyżelewicz, D.; Hodurek, P. Centrifugal partition chromatography—a review of recent applications and some classic references. J. Sep. Sci. 2017, 40, 1597–1609. [Google Scholar] [CrossRef]
- Song, H.; Lin, J.; Zhu, X.; Chen, Q. Developments in high-speed countercurrent chromatography and its applications in the separation of terpenoids and saponins. J. Sep. Sci. 2016, 39, 1574–1591. [Google Scholar] [CrossRef]
- Skalicka-Woźniak, K.; Garrard, I. Counter-current chromatography for the separation of terpenoids: A comprehensive review with respect to the solvent systems employed. Phytochem. Rev. 2014, 13, 547–572. [Google Scholar] [CrossRef] [Green Version]
- Pauli, G.F.; Pro, S.M.; Friesen, J.B. Countercurrent separation of natural products. J. Nat. Prod. 2008, 71, 1489–1508. [Google Scholar] [CrossRef]
- Friesen, J.B.; McAlpine, J.B.; Chen, S.-N.; Pauli, G.F. Countercurrent separation of natural products: An update. J. Nat. Prod. 2015, 78, 1765–1796. [Google Scholar] [CrossRef] [Green Version]
- Hammerschick, T.; Wagner, T.; Vetter, W. Isolation of saturated alkylresorcinols from rye grains by countercurrent chromatography. J. Sep. Sci. 2021, 44, 1904–1912. [Google Scholar] [CrossRef]
- Hammerschick, T.; Vetter, W. Online hyphenation of centrifugal partition chromatography with countercurrent chromatography (CPC-CCC) and its application to the separation of saturated alkylresorcinols. Anal. Bioanal. Chem. 2022, 414, 5043–5051. [Google Scholar] [CrossRef]
- Navruz-Varli, S.; Sanlier, N. Nutritional and health benefits of quinoa (Chenopodium quinoa Willd.). J. Cereal Sci. 2016, 69, 371–376. [Google Scholar] [CrossRef]
- Müller, M.; Hammann, S.; Vetter, W. Countercurrent chromatographic isolation and purification of 11′-α-tocomonoenol from the vitamin E extract of palm oil. Food Chem. 2018, 256, 327–332. [Google Scholar] [CrossRef] [Green Version]
- Marchal, L.; Intes, O.; Foucault, A.; Legrand, J.; Nuzillard, J.-M.; Renault, J.-H. Rational improvement of centrifugal partition chromatographic settings for the production of 5-n-alkylresorcinols from wheat bran lipid extract. J. Chromatogr. A 2003, 1005, 51–62. [Google Scholar] [CrossRef]
- Przybylski, R.; Chauhan, G.; Eskin, N. Characterization of quinoa (Chenopodium quinoa) lipids. Food Chem. 1994, 51, 187–192. [Google Scholar] [CrossRef]
- Berthod, A.; Ruiz-Angel, M.J.; Carda-Broch, S. Elution-extrusion countercurrent chromatography. Use of the liquid nature of the stationary phase to extend the hydrophobicity window. Anal. Chem. 2003, 75, 5886–5894. [Google Scholar] [CrossRef]
- Kaneda, T. Iso- and anteiso-fatty acids in bacteria: Biosynthesis, function, and taxonomic significance. Microbiol. Rev. 1991, 55, 288–302. [Google Scholar] [CrossRef]
- Hirosuke, O.; Noriyasu, Y.; Junichi, N.; Isao, C. Precursor role of branched-chain amino acids in the biosynthesis of iso and anteiso fatty acids in rat skin. BBA—Lipids Lipid Metab. 1994, 1214, 279–287. [Google Scholar] [CrossRef]
- Thurnhofer, S.; Hottinger, G.; Vetter, W. Enantioselective determination of anteiso fatty acids in food samples. Anal. Chem. 2007, 79, 4696–4701. [Google Scholar] [CrossRef] [PubMed]
- Eibler, D.; Abdurahman, H.; Ruoff, T.; Kaffarnik, S.; Steingass, H.; Vetter, W. Unexpected formation of low amounts of (R)-configurated anteiso-fatty acids in rumen fluid experiments. PLoS ONE 2017, 12, e0170788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauff, S.; Hottinger, G.; Vetter, W. Enantioselective analysis of chiral anteiso fatty acids in the polar and neutral lipids of food. Lipids 2010, 45, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Murić, M.; Glanz, L.; Vetter, W. Improving the resolution of overlapping peaks by heartcut two-dimensional countercurrent chromatography with the same solvent system in both dimensions. J. Chromatogr. A 2019, 1596, 142–151. [Google Scholar] [CrossRef]
- Englert, M.; Brown, L.; Vetter, W. Heart-cut two-dimensional countercurrent chromatography with a single instrument. Anal. Chem. 2015, 87, 10172–10177. [Google Scholar] [CrossRef]
- Rüttler, F.; Hammerschick, T.; Schlag, S.; Vetter, W. Isolation of lanosterol and dihydrolanosterol from the unsaponifiable matter of lanolin by urea complexation and countercurrent chromatography in heart–cut recycling mode. J. Chromatogr. B 2022, 1210, 123470. [Google Scholar] [CrossRef]
- Kazantzoglou, G.; Magiatis, P.; Kalpoutzakis, E.; Skaltsounis, A.-L. Polygonophenone, the first MEM-substituted natural product, from Polygonum maritimum. J. Nat. Prod. 2009, 72, 187–189. [Google Scholar] [CrossRef]
- Friesen, J.B.; Pauli, G.F. Rational development of solvent system families in counter-current chromatography. J. Chromatogr. A 2007, 1151, 51–59. [Google Scholar] [CrossRef]
- Knödler, M.; Kaiser, A.; Carle, R.; Schieber, A. Profiling of alk(en)ylresorcinols in cereals by HPLC-DAD-APcI-MSn. Anal. Bioanal. Chem. 2008, 391, 221–228. [Google Scholar] [CrossRef]
- Kowalska, I.; Jędrejek, D. Benzoxazinoid and alkylresorcinol content, and their antioxidant potential, in a grain of spring and winter wheat cultivated under different production systems. J. Cereal Sci. 2020, 95, 103063. [Google Scholar] [CrossRef]








| Alkylresorcinol (AR) a | m/z [M]+ | GC tR [min] | Contribution to ΣARs | K Value b | Methylalkylresorcinol (mAR) c | m/z [M]+ | GC tR [min] | Contribution to ΣARs | K Value b |
|---|---|---|---|---|---|---|---|---|---|
| Saturated | Saturated | ||||||||
| AR16:0 | 478 | 22.94 | tr | 0.31 | |||||
| AR17:0 * | 492 | 24.57 | tr | 0.40 | mAR17:0 * | 506 | 25.50 | tr | 0.54 |
| AR18:0 * | 506 | 26.31 | tr | 0.51 | mAR18:0 | 520 | 27.28 | tr | 0.65 |
| AR19:0 * | 520 | 28.20 | 3.4% | 0.65 | mAR19:0 * | 534 | 29.28 | 1.0% | 0.82 |
| AR20:0 * | 534 | 30.14 | 0.3% | 0.82 | mAR20:0 | 548 | 31.27 | 0.1% | 1.07 |
| AR21:0 * | 548 | 32.11 | 11.0% | 1.06 | mAR21:0 * | 562 | 33.27 | 2.2% | 1.37 |
| AR22:0 * | 562 | 34.09 | 0.5% | 1.33 | mAR22:0 | 576 | 35.26 | 0.1% | 1.75 |
| AR23:0 * | 576 | 36.10 | 2.5% | 1.71 | mAR23:0 * | 590 | 37.28 | 0.4% | 2.16 |
| AR24:0 * | 590 | 38.07 | tr | 2.11 | mAR24:0 | 604 | 39.27 | tr | 2.78 |
| AR25:0 * | 604 | 40.04 | 0.3% | 2.61 | mAR25:0 | 618 | 41.26 | tr | d |
| AR26:0 * | 618 | 42.00 | tr | d | |||||
| AR15:0-i | 464 | 21.47 | tr | 0.28 | mAR15:0-i | 478 | 22.16 | tr | 0.33 |
| AR17:0-i * | 492 | 23.93 | tr | 0.39 | mAR17:0-i | 506 | 24.81 | tr | 0.51 |
| AR19:0-i * | 520 | 27.47 | 2.3% | 0.61 | mAR19:0-i * | 534 | 28.55 | 1.4% | 0.78 |
| AR20:0-i | 534 | 29.36 | tr | 0.75 | mAR20:0-i | 548 | 30.51 | tr | 0.96 |
| AR21:0-i * | 548 | 31.36 | 14.3% | 0.99 | mAR21:0-i * | 562 | 32.52 | 5.2% | 1.30 |
| AR22:0-i * | 562 | 33.32 | tr | 1.23 | mAR22:0-i | 576 | 34.51 | tr | 1.61 |
| AR23:0-i * | 576 | 35.34 | 9.5% | 1.61 | mAR23:0-i * | 590 | 36.54 | 2.4% | 2.06 |
| AR24:0-i * | 590 | 37.29 | tr | 2.02 | mAR24:0-i | 604 | 38.54 | tr | 2.61 |
| AR25:0-i * | 604 | 39.32 | 1.5% | 2.51 | mAR25:0-i * | 618 | 40.52 | tr | d |
| AR26:0-i | 618 | 41.27 | tr | d | |||||
| AR27:0-i | 632 | 43.38 | tr | d | |||||
| AR16:0-a | 478 | 22.55 | tr | 0.30 | |||||
| AR18:0-a * | 506 | 25.83 | 0.2% | 0.47 | mAR18:0-a * | 520 | 26.85 | 0.1% | 0.61 |
| AR19:0-a * | 520 | 27.59 | tr | 0.61 | mAR19:0-a * | 534 | 28.65 | tr | 0.79 |
| AR20:0-a * | 534 | 29.62 | 9.0% | 0.75 | mAR20:0-a * | 548 | 30.76 | 3.9% | 0.96 |
| AR21:0-a * | 548 | 31.54 | tr | 0.99 | mAR21:0-a * | 562 | 32.74 | tr | 1.30 |
| AR22:0-a * | 562 | 33.59 | 12.4% | 1.23 | mAR22:0-a * | 576 | 34.78 | 4.9% | 1.61 |
| AR23:0-a * | 576 | 35.51 | tr | 1.58 | mAR23:0-a * | 590 | 36.78 | tr | 2.06 |
| AR24:0-a * | 590 | 37.58 | 4.2% | 1.96 | mAR24:0-a * | 604 | 38.79 | 0.9% | 2.51 |
| AR25:0-a | 604 | 39.57 | tr | 2.44 | |||||
| AR26:0-a * | 618 | 41.52 | 0.2% | d | mAR26:0-a * | 632 | 42.75 | tr | d |
| Monoenoic | Monoenoic | ||||||||
| mAR17:1 | 504 | 25.05 | tr | 0.33 | |||||
| AR19:1 * | 518 | 27.71, 27.89 | 0.1% | 0.44 | mAR19:1 | 532 | 28.76, 28.96 | tr | 0.54 |
| AR20:1 | 532 | 29.72 | tr | 0.54 | |||||
| AR21:1 * | 546 | 31.62, 31.81 | 2.2% | 0.68 | mAR21:1 * | 560 | 32.83, 33.00 | 0.5% | 0.85 |
| AR22:1 * | 560 | 33.65 | tr | 0.82 | |||||
| AR23:1 * | 574 | 35.70, 35.84 | 0.5% | 1.06 | mAR23:1 | 588 | 36.85, 37.05 | 0.1% | 1.40 |
| AR25:1 | 602 | 39.68 | tr | 1.71 | mAR25:1 | 616 | 40.91 | tr | 2.16 |
| AR27:1 | 630 | 43.87 | tr | 2.68 | |||||
| AR21:1-i | 546 | 30.73, 30.88 | tr | 0.61 | mAR21:1-i | 560 | 32.06 | tr | 0.78 |
| AR20:1-a | 532 | 29.07 | tr | 0.54 | |||||
| AR22:1-a | 560 | 33.06 | 0.2% | 0.75 | mAR22:1-a * | 574 | 34.28 | 0.1% | 1.02 |
| AR24:1-a | 588 | 37.12 | tr | 1.23 | mAR24:1-a | 602 | 38.30 | tr | 1.58 |
| AR26:1-a | 616 | 41.10 | tr | 1.92 | |||||
| Dienoic | Dienoic | ||||||||
| AR17:2 | 488 | 27.55 | tr | 0.30 | |||||
| mAR19:2 | 530 | 28.74 | tr | 0.40 | |||||
| AR21:2 * | 544 | 31.60 | 0.5% | 0.33 | mAR21:2 | 558 | 32.79 | 0.1% | 0.61 |
| AR22:2 | 558 | 33.47 | tr | 0.47 | |||||
| AR23:2 * | 572 | 35.66 | 0.1% | 0.75 | mAR23:2 | 586 | 36.89 | tr | 0.99 |
| AR25:2 | 600 | 39.63 | tr | 1.16 | |||||
| Trienoic | Trienoic | ||||||||
| AR21:3 | 542 | 31.82 | tr | 0.33 | |||||
| Keto group | Keto group | ||||||||
| AR19:0 oxo | 534 | 30.51 | tr | 0.27 | mAR19:0 oxo | 548 | 31.46 | tr | 0.32 |
| AR21:0 oxo | 562 | 34.54 | tr | 0.33 | |||||
| AR23:0 oxo | 590 | 38.53 | tr | 0.51 | |||||
| mAR19:0-i oxo | 548 | 30.80 | tr | 0.31 | |||||
| AR21:0-i oxo | 562 | 33.64 | tr | 0.33 | mAR21:0-i oxo | 576 | 34.78 | tr | 0.51 |
| AR23:0-i oxo | 590 | 37.75 | tr | 0.51 | |||||
| AR25:0-i oxo | 618 | 41.76 | tr | 0.82 | |||||
| AR20:0-a oxo | 548 | 31.94 | tr | 0.30 | mAR20:0-a oxo | 562 | 33.04 | tr | 0.37 |
| AR22:0-a oxo | 576 | 36.00 | tr | 0.37 | mAR22:0-a oxo | 590 | 37.12 | tr | 0.58 |
| AR24:0-a oxo | 604 | 39.98 | tr | 0.58 | |||||
| AR23:1 oxo | 588 | 38.1 | tr | 0.33 |
| 1st Dimension | 2nd Dimension | ||
|---|---|---|---|
| Heart-Cut | Performed a (Intended b) Transfer Range [mL] | Targeted AR (Purities) | Purities of the Target AR in the Pooled Heart-Cut Fractions for SIC c |
| AT→H, HC 1 | 132–151 (123–151) | mAR23:0-i (22%) AR24:0-a (38%) | mAR23:0-i d AR24:0-a (58%, 144–164 mL) |
| AT→H, HC 2 | 151–173 (151–182) | mAR22:0-a (26%) AR23:0-i (37%) | mAR22:0-a (45%, 153–174 mL) AR23:0-i d |
| AT→H, HC 3 | 188–212 (197–221) | mAR21:0-i (18%) AR22:0-a (58%) | mAR21:0-i (25%, 307–328 mL) AR22:0-a (60%, 230–251 mL) |
| AT→H, HC 4 | 212–289 (221–280) | mAR20:0-a (15%) AR21:0-i (48%) | mAR20:0-a (22%, 244–307 mL) AR21:0-i d |
| BH→T, HC 1 | 134–156 (136–157) | mAR18:0-a (2%) AR19:0-i (45%) | mAR18:0-a (3%, 134.5–169.5) AR19:0-i d |
| BH→T, HC 2 | 172–204.5 (172–205) | mAR19:0-i (11%) AR20:0-a (65%) | mAR19:0-i (11%, 239.5–269.5 mL) AR20:0-a (88%, 239.5–269.5 mL) |
| Isolated (m)AR | Mass [mg] | Purity [%] | CCC Elution Volumes Used for SIC |
|---|---|---|---|
| mAR18:0-a | 0.6 | >97% | BH→T, HC 1 + 2: 134.5–169.5 mL |
| mAR19:0-i | 0.9 | >98% | BH→T, HC 1 + 2: 239.5–269.5 mL |
| mAR20:0-a | 15.5 | >97% | AT→H, HC 2 + 3: 349–426 mL, HC 4: 244–307 mL |
| mAR21:0-i | 1.7 | >93% | AT→H, HC 2 + 3: 307–328 mL, HC 4: 202–223 mL |
| mAR22:0-a | 15.3 | >96% | AT→H, HC 1: 184–209 mL, HC 2 + 3: 153–174 mL |
| mAR24:0-a | 2.4 | >95% | AT→H, 104–114 mL |
| AR20:0-a | 7.5 | >96% | BH→T, HC 1 + 2, 239.5–269.5 mL |
| AR21:0-i | 3.0 | >90% | AT→H, HC 1: 321–342 mL; BH→T, HC 1 + 2: 309.5–339.5 mL |
| AR22:0-a | 13.0 | >95% | AT→H, HC 2 + 3: 230–251 mL, 279–293 mL, HC 4: 188–216 mL |
| AR24:0-a | 3.8 | >95% | AT→H, HC 1: 144–164 mL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hammerschick, T.; Vetter, W. Profiling and Isolation of Ten Rare Branched-Chain Alkylresorcinols in Quinoa. Molecules 2023, 28, 5220. https://doi.org/10.3390/molecules28135220
Hammerschick T, Vetter W. Profiling and Isolation of Ten Rare Branched-Chain Alkylresorcinols in Quinoa. Molecules. 2023; 28(13):5220. https://doi.org/10.3390/molecules28135220
Chicago/Turabian StyleHammerschick, Tim, and Walter Vetter. 2023. "Profiling and Isolation of Ten Rare Branched-Chain Alkylresorcinols in Quinoa" Molecules 28, no. 13: 5220. https://doi.org/10.3390/molecules28135220
APA StyleHammerschick, T., & Vetter, W. (2023). Profiling and Isolation of Ten Rare Branched-Chain Alkylresorcinols in Quinoa. Molecules, 28(13), 5220. https://doi.org/10.3390/molecules28135220