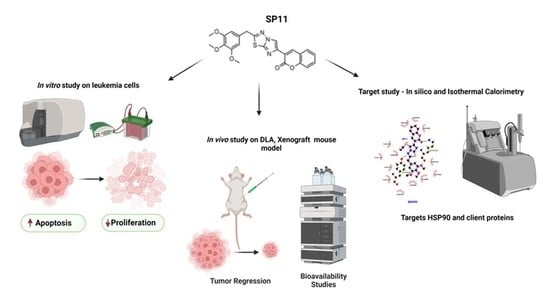
A Coumarin–Imidazothiadiazole Derivative, SP11 Abrogates Tumor Growth by Targeting HSP90 and Its Client Proteins
Abstract
:1. Introduction
2. Results
2.1. Synthesis
2.2. SP11 Induces Cytotoxicity in Various Leukemia Cell Lines with Least Effect on Normal Cells
2.3. SP11 Treatment Induces Apoptosis Rather Than Necrosis in Leukemic Cells
2.4. SP11 Induces Tumor Regression in DLA Induced Allograft Mouse Tumor Model
2.5. SP11 Reduced Tumor Burden with Minimal Toxicity and Enhanced Bioavailability
2.6. SP11 Binds to the C-Terminal of HSP90
2.7. Isothermal Calorimetry
2.8. SP11 Regulates Expression of HSP90 Client Proteins
2.9. SP11 Induces Tumor Regression in a Xenograft Mouse Model and Regulates Expression of HSP90 Client Proteins
3. Discussion
4. Material and Methods
4.1. Synthesis of 3-(2-(3,4,5-Trimethoxybenzyl)imidazo [2,1-b][1,3,4]thiadiazol-6-yl)-2H-chromen-2-one) (5)
4.2. Synthesis of 3-(5-Thiocyanato-2-(3,4,5-trimethoxybenzyl)imidazo [2,1-b][1,3,4]thiadiazol-6-yl-2H-chromen-2-one 6 (SP11)
4.3. Reagents, Chemicals, and Apparatus
4.4. Cell Culture
4.5. MTT Assay
4.6. LDH Assay
4.7. Apoptosis Assay
4.8. Immunoblotting
4.9. In-Cell Western
4.10. Dalton’s Lymphoma Tumor Model
4.11. Drug Toxicity and Side-Effect Assessment on ST09 Treatment
4.12. Histological Analysis of Tumor Tissues
4.13. Collagen Staining
4.14. SP11 Bioavailability Studies
4.14.1. Chromatographic Conditions
4.14.2. Sample Preparation
4.14.3. HPLC Analysis
4.15. Leukemia Xenograft Study
4.16. Docking Studies
4.17. Cloning, Expression, and Purification of HSP90 C-Terminal Domain (CTD) and N-Terminal Domain (NTD)
4.18. Isothermal Calorimetry
4.19. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, L.; You, Q.-D.; Xu, X.-L. Heat Shock Protein 90 Inhibitors: An Update on Achievements, Challenges, and Future Directions. J. Med. Chem. 2020, 63, 1798–1822. [Google Scholar] [CrossRef] [PubMed]
- Zuehlke, A.D.; Moses, M.A.; Neckers, L. Heat Shock Protein 90: Its Inhibition and Function. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20160527. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.; Lee, H.W.; Lee, E.H.; Park, M.-I.; Lee, J.S.; Kim, M.-S.; Kim, K.; Roh, M.S.; Pak, M.G.; Oh, J.E.; et al. Differential Expression of HSP90 Isoforms and Their Correlations with Clinicopathologic Factors in Patients with Colorectal Cancer. Int. J. Clin. Exp. Pathol. 2019, 12, 978–986. [Google Scholar]
- Koga, F.; Xu, W.; Karpova, T.S.; McNally, J.G.; Baron, R.; Neckers, L. Hsp90 Inhibition Transiently Activates Src Kinase and Promotes Src-Dependent Akt and Erk Activation. Proc. Natl. Acad. Sci. USA 2006, 103, 11318–11322. [Google Scholar] [CrossRef]
- Yang, H.; Lee, M.-H.; Park, I.; Jeon, H.; Choi, J.; Seo, S.; Kim, S.-W.; Koh, G.Y.; Park, K.-S.; Lee, D.H. HSP90 Inhibitor (NVP-AUY922) Enhances the Anti-Cancer Effect of BCL-2 Inhibitor (ABT-737) in Small Cell Lung Cancer Expressing BCL-2. Cancer Lett. 2017, 411, 19–26. [Google Scholar] [CrossRef]
- Sato, S.; Fujita, N.; Tsuruo, T. Modulation of Akt Kinase Activity by Binding to Hsp90. Proc. Natl. Acad. Sci. USA 2000, 97, 10832–10837. [Google Scholar] [CrossRef]
- Trisciuoglio, D.; Gabellini, C.; Desideri, M.; Ziparo, E.; Zupi, G.; Del Bufalo, D. Bcl-2 Regulates HIF-1α Protein Stabilization in Hypoxic Melanoma Cells via the Molecular Chaperone HSP90. PLoS ONE 2010, 5, e11772. [Google Scholar] [CrossRef]
- Tu, R.-H.; Li, Q.-J.; Huang, Z.; He, Y.; Meng, J.-J.; Zheng, H.-L.; Zeng, Z.-Y.; Zhong, G.-Q. Novel Functional Role of Heat Shock Protein 90 in Mitochondrial Connexin 43-Mediated Hypoxic Postconditioning. Cell. Physiol. Biochem. 2017, 44, 982–997. [Google Scholar] [CrossRef] [Green Version]
- Hallett, S.T.; Pastok, M.W.; Morgan, R.M.L.; Wittner, A.; Blundell, K.L.I.M.; Felletar, I.; Wedge, S.R.; Prodromou, C.; Noble, M.E.M.; Pearl, L.H.; et al. Differential Regulation of G1 CDK Complexes by the Hsp90-Cdc37 Chaperone System. Cell Rep. 2017, 21, 1386–1398. [Google Scholar] [CrossRef]
- Chakraborty, A.; Edkins, A.L. HSP90 as a Regulator of Extracellular Matrix Dynamics. Biochem. Soc. Trans. 2021, 49, 2611–2625. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.C.; O’Hagan, K.L.; Kenyon, A.; Dhanani, K.C.H.; Prinsloo, E.; Edkins, A.L. Hsp90 Binds Directly to Fibronectin (FN) and Inhibition Reduces the Extracellular Fibronectin Matrix in Breast Cancer Cells. PLoS ONE 2014, 9, e86842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The Extracellular Matrix at a Glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubow, K.E.; Vukmirovic, R.; Zhe, L.; Klotzsch, E.; Smith, M.L.; Gourdon, D.; Luna, S.; Vogel, V. Mechanical Forces Regulate the Interactions of Fibronectin and Collagen I in Extracellular Matrix. Nat. Commun. 2015, 6, 8026. [Google Scholar] [CrossRef] [Green Version]
- Gorska, M. Geldanamycin and Its Derivatives as Hsp90 Inhibitors. Front. Biosci. 2012, 17, 2269. [Google Scholar] [CrossRef] [Green Version]
- Le Brazidec, J.Y.; Kamal, A.; Busch, D.; Thao, L.; Zhang, L.; Timony, G.; Grecko, R.; Trent, K.; Lough, R.; Salazar, T.; et al. Synthesis and biological evaluation of a new class of geldanamycin derivatives as potent inhibitors of Hsp90. J. Med. Chem. 2004, 47, 3865–3873. [Google Scholar] [CrossRef]
- Miyata, Y. Hsp90 Inhibitor Geldanamycin and Its Derivatives as Novel Cancer Chemotherapeutic Agents. Curr. Pharm. Des. 2005, 11, 1131–1138. [Google Scholar] [CrossRef]
- Villa, R.; Folini, M.; Porta, C.D.; Valentini, A.; Pennati, M.; Daidone, M.G.; Zaffaroni, N. Inhibition of Telomerase Activity by Geldanamycin and 17-Allylamino, 17-Demethoxygeldanamycin in Human Melanoma Cells. Carcinogenesis 2003, 24, 851–859. [Google Scholar] [CrossRef] [Green Version]
- Whitesell, L.; Lindquist, S.L. HSP90 and the Chaperoning of Cancer. Nat. Rev. Cancer 2005, 5, 761–772. [Google Scholar] [CrossRef]
- Marcu, M.G.; Schulte, T.W.; Neckers, L. Novobiocin and Related Coumarins and Depletion of Heat Shock Protein 90-Dependent Signaling Proteins. J. Natl. Cancer Inst. 2000, 92, 242–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allan, R.K.; Mok, D.; Ward, B.K.; Ratajczak, T. Modulation of Chaperone Function and Cochaperone Interaction by Novobiocin in the C-Terminal Domain of Hsp90: Evidence That Coumarin Antibiotics Disrupt Hsp90 Dimerization. J. Biol. Chem. 2006, 281, 7161–7171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazenby, M.; Hills, R.; Burnett, A.K.; Zabkiewicz, J. The HSP90 Inhibitor Ganetespib: A Potential Effective Agent for Acute Myeloid Leukemia in Combination with Cytarabine. Leuk. Res. 2015, 39, 617–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Saini, N.; Parris, A.B.; Zhao, M.; Yang, X. Ganetespib Induces G2/M Cell Cycle Arrest and Apoptosis in Gastric Cancer Cells through Targeting of Receptor Tyrosine Kinase Signaling. Int. J. Oncol. 2017, 51, 967–974. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Saini, N.; Howard, E.W.; Parris, A.B.; Ma, Z.; Zhao, Q.; Zhao, M.; Liu, B.; Edgerton, S.M.; Thor, A.D.; et al. Ganetespib Targets Multiple Levels of the Receptor Tyrosine Kinase Signaling Cascade and Preferentially Inhibits ErbB2-Overexpressing Breast Cancer Cells. Sci. Rep. 2018, 8, 6829. [Google Scholar] [CrossRef] [Green Version]
- Fennell, D.; Antonov, A.; Martins, M.L.; Popat, S.; Ramalingam, S.S.; Spicer, J.; Vukovic, V.M.; El-Hariry, I.; Reichert, V.; Rosell, R. Abstract 4657: Evaluation of Genomic Profiling in the GALAXY-1 (NCT01348126), a Randomized Phase 2b Study of Ganetespib in Combination with Docetaxel versus Docetaxel Alone as Second Line Therapy in Patients with Advanced NSCLC. Clin. Res. (Exclud. Clin. Trials) 2014, 74, 4657. [Google Scholar] [CrossRef]
- Zhao, H.; Yan, B.; Peterson, L.B.; Blagg, B.S.J. 3-Arylcoumarin Derivatives Manifest Anti-Proliferative Activity through Hsp90 Inhibition. ACS Med. Chem. Lett. 2012, 3, 327–331. [Google Scholar] [CrossRef]
- Zhao, H.; Brandt, G.E.; Galam, L.; Matts, R.L.; Blagg, B.S.J. Identification and Initial SAR of Silybin: An Hsp90 Inhibitor. Bioorg. Med. Chem. Lett. 2011, 21, 2659–2664. [Google Scholar] [CrossRef]
- Shelton, S.N.; Shawgo, M.E.; Matthews, S.B.; Lu, Y.; Donnelly, A.C.; Szabla, K.; Tanol, M.; Vielhauer, G.A.; Rajewski, R.A.; Matts, R.L.; et al. KU135, a Novel Novobiocin-Derived C-Terminal Inhibitor of the 90-kDa Heat Shock Protein, Exerts Potent Antiproliferative Effects in Human Leukemic Cells. Mol. Pharmacol. 2009, 76, 1314–1322. [Google Scholar] [CrossRef] [Green Version]
- Samadi, A.K.; Zhang, X.; Mukerji, R.; Donnelly, A.C.; Blagg, B.S.; Cohen, M.S. A Novel C-Terminal HSP90 Inhibitor KU135 Induces Apoptosis and Cell Cycle Arrest in Melanoma Cells. Cancer Lett. 2011, 312, 158–167. [Google Scholar] [CrossRef] [Green Version]
- Cikotiene, I.; Kazlauskas, E.; Matuliene, J.; Michailoviene, V.; Torresan, J.; Jachno, J.; Matulis, D. 5-Aryl-4-(5-Substituted-2,4-Dihydroxyphenyl)-1,2,3-Thiadiazoles as Inhibitors of Hsp90 Chaperone. Bioorg. Med. Chem. Lett. 2009, 19, 1089–1092. [Google Scholar] [CrossRef]
- Venugopala, K.N.; Rashmi, V.; Odhav, B. Review on Natural Coumarin Lead Compounds for Their Pharmacological Activity. BioMed Res. Int. 2013, 2013, 963248. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Xu, J.; Liu, Y.; Zeng, Y.; Wu, G. A Review on Anti-Tumor Mechanisms of Coumarins. Front. Oncol. 2020, 10, 592853. [Google Scholar] [CrossRef]
- Szeliga, M. Thiadiazole Derivatives as Anticancer Agents. Pharmacol. Rep. 2020, 72, 1079–1100. [Google Scholar] [CrossRef]
- Hurwitz, R.; Hozier, J.; Lebien, T.; Minowada, J.; Gajl-Peczalska, K.; Kubonishi, I.; Kersey, J. Bromination of 3-Acetocoumarin.-B Phenotype. Int. J. Cancer 1979, 23, 174–180. [Google Scholar] [CrossRef]
- Medh, R.D.; Scott Webb, M.; Miller, A.L.; Johnson, B.H.; Fofanov, Y.; Li, T.; Wood, T.G.; Luxon, B.A.; Brad Thompson, E. Gene Expression Profile of Human Lymphoid CEM Cells Sensitive and Resistant to Glucocorticoid-Evoked Apoptosis. Genomics 2003, 81, 543. [Google Scholar] [CrossRef] [Green Version]
- Klein, E.; Ben-Bassat, H.; Neumann, H.; Ralph, P.; Zeuthen, J.; Polliack, A.; Vánky, F. Properties of the K562 Cell Line, Derived from a Patient with Chronic Myeloid Leukemia. Int. J. Cancer 1976, 18, 421–431. [Google Scholar] [CrossRef]
- Kayibanda, B.; Rosenfeld, C.; Goutner, A.; Bornkamm, G.W. A New Lymphoid Cell Line, Reh 6, with Characteristics of Non-T and Non-B Cells, Lacking the Epstein-Barr Virus Genome and Derived from Human Acute Lymphoblastic Leukemia. Intervirology 1978, 9, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, J.M.; Gonzalez-Sarmiento, R.; Arthur, D.C.; Wilkowski, C.W.; Streifel, B.J.; Kersey, J.H. Immunophenotypic and Cytogenetic Analysis of Molt-3 and Molt-4: Human T-Lymphoid Cell Lines with Rearrangement of Chromosome 7. Blood 1988, 72, 1755–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crowley, L.C.; Marfell, B.J.; Scott, A.P.; Waterhouse, N.J. Quantitation of Apoptosis and Necrosis by Annexin V Binding, Propidium Iodide Uptake, and Flow Cytometry. Cold Spring Harb. Protoc. 2016, 2016, pdb-prot087288. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.R. Dalton’s Lymphoma as a Murine Model for Understanding the Progression and Development of T-Cell Lymphoma and Its Role in Drug Discovery. Int. J. Immunother. Cancer Res. 2017, 3, 001–006. [Google Scholar] [CrossRef]
- Prince, T.; Sun, L.; Matts, R.L. Cdk2: A Genuine Protein Kinase Client of Hsp90 and Cdc37. Biochemistry 2005, 44, 15287–15295. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Miguel, T.; Gajate, C.; González-Camacho, F.; Mollinedo, F. Proapoptotic Role of Hsp90 by Its Interaction with c-Jun N-Terminal Kinase in Lipid Rafts in Edelfosine-Mediated Antileukemic Therapy. Oncogene 2008, 27, 1779–1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Wang, X.; Cao, C.; Wang, X.; Liang, S.; Peng, C.; Fu, L.; He, G. Inhibition of HSP90 Sensitizes a Novel Raf/ERK Dual Inhibitor CY-9d in Triple-Negative Breast Cancer Cells. Oncotarget 2017, 8, 104193–104205. [Google Scholar] [CrossRef] [Green Version]
- Ota, A.; Zhang, J.; Ping, P.; Han, J.; Wang, Y. Specific Regulation of Noncanonical p38alpha Activation by Hsp90-Cdc37 Chaperone Complex in Cardiomyocyte. Circ. Res. 2010, 106, 1404–1412. [Google Scholar] [CrossRef]
- Sun, P.; Wang, Y.; Gao, T.; Li, K.; Zheng, D.; Liu, A.; Ni, Y. Hsp90 Modulates Human Sperm Capacitation via the Erk1/2 and p38 MAPK Signaling Pathways. Reprod. Biol. Endocrinol. 2021, 19, 39. [Google Scholar] [CrossRef]
- Burnett, A.; Wetzler, M.; Löwenberg, B. Therapeutic Advances in Acute Myeloid Leukemia. J. Clin. Oncol. 2011, 29, 487–494. [Google Scholar] [CrossRef] [Green Version]
- Beebe, K.; Mollapour, M.; Scroggins, B.; Prodromou, C.; Xu, W.; Tokita, M.; Taldone, T.; Pullen, L.; Zierer, B.K.; Lee, M.-J.; et al. Posttranslational Modification and Conformational State of Heat Shock Protein 90 Differentially Affect Binding of Chemically Diverse Small Molecule Inhibitors. Oncotarget 2013, 4, 1065–1074. [Google Scholar] [CrossRef] [Green Version]
- Moulick, K.; Ahn, J.H.; Zong, H.; Rodina, A.; Cerchietti, L.; Gomes DaGama, E.M.; Caldas-Lopes, E.; Beebe, K.; Perna, F.; Hatzi, K.; et al. Affinity-Based Proteomics Reveal Cancer-Specific Networks Coordinated by Hsp90. Nat. Chem. Biol. 2011, 7, 818–826. [Google Scholar] [CrossRef] [Green Version]
- Jhaveri, K.; Ochiana, S.O.; Dunphy, M.P.; Gerecitano, J.F.; Corben, A.D.; Peter, R.I.; Janjigian, Y.Y.; Gomes-DaGama, E.M.; Koren, J.; Modi, S.; et al. Heat Shock Protein 90 Inhibitors in the Treatment of Cancer: Current Status and Future Directions. Expert Opin. Investig. Drugs 2014, 23, 611–628. [Google Scholar] [CrossRef] [Green Version]
- Lancet, J.E.; Gojo, I.; Burton, M.; Quinn, M.; Tighe, S.M.; Kersey, K.; Zhong, Z.; Albitar, M.X.; Bhalla, K.; Hannah, A.L.; et al. Phase I Study of the Heat Shock Protein 90 Inhibitor Alvespimycin (KOS-1022, 17-DMAG) Administered Intravenously Twice Weekly to Patients with Acute Myeloid Leukemia. Leukemia 2010, 24, 699–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zong, H.; Gozman, A.; Caldas-Lopes, E.; Taldone, T.; Sturgill, E.; Brennan, S.; Ochiana, S.O.; Gomes-DaGama, E.M.; Sen, S.; Rodina, A.; et al. A Hyperactive Signalosome in Acute Myeloid Leukemia Drives Addiction to a Tumor-Specific Hsp90 Species. Cell Rep. 2015, 13, 2159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Effects of Green Synthesised Silver Nanoparticles (ST06-AgNPs) Using Curcumin Derivative (ST06) on Human Cervical Cancer Cells (HeLa) in Vitro and EAC Tumor Bearing Mice Models [Corrigendum]. Int. J. Nanomed. 2019, 14, 6133. [CrossRef] [PubMed] [Green Version]
- Cerchietti, L.C.; Lopes, E.C.; Yang, S.N.; Hatzi, K.; Bunting, K.L.; Tsikitas, L.A.; Mallik, A.; Robles, A.I.; Walling, J.; Varticovski, L.; et al. A Purine Scaffold Hsp90 Inhibitor Destabilizes BCL-6 and Has Specific Antitumor Activity in BCL-6-Dependent B Cell Lymphomas. Nat. Med. 2009, 15, 1369–1376. [Google Scholar] [CrossRef]
- Chen, C.; Zhuang, Y.; Chen, X.; Chen, X.; Li, D.; Fan, Y.; Xu, J.; Chen, Y.; Wu, L. Hsp90 N- and C-Terminal Double Inhibition Synergistically Suppresses Bcr-Abl-Positive Human Leukemia Cells. Oncotarget 2017, 8, 10025. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; McAlpine, S.R. N-Terminal and C-Terminal Modulation of Hsp90 Produce Dissimilar Phenotypes. Chem. Commun. 2015, 51, 1410–1413. [Google Scholar] [CrossRef] [Green Version]
- Trepel, J.; Mollapour, M.; Giaccone, G.; Neckers, L. Targeting the Dynamic HSP90 Complex in Cancer. Nat. Rev. Cancer 2010, 10, 537–549. [Google Scholar] [CrossRef] [Green Version]
- Donnelly, A.; Blagg, B.S. Novobiocin and Additional Inhibitors of the Hsp90 C-Terminal Nucleotide-Binding Pocket. Curr. Med. Chem. 2008, 15, 2702–2717. [Google Scholar] [CrossRef] [Green Version]
- Su, H.; Xu, Y. Application of ITC-Based Characterization of Thermodynamic and Kinetic Association of Ligands With Proteins in Drug Design. Front. Pharmacol. 2018, 9, 1133. [Google Scholar] [CrossRef]
- Nirgude, S.; Mahadeva, R.; Koroth, J.; Kumar, S.; Kumar, K.S.S.; Gopalakrishnan, V.; Karki, S.S.S.; Choudhary, B. ST09, A Novel Curcumin Derivative, Blocks Cell Migration by Inhibiting Matrix Metalloproteases in Breast Cancer Cells and Inhibits Tumor Progression in EAC Mouse Tumor Models. Molecules 2020, 25, 4499. [Google Scholar] [CrossRef]
- Koroth, J.; Nirgude, S.; Tiwari, S.; Gopalakrishnan, V.; Mahadeva, R.; Kumar, S.; Karki, S.S.; Choudhary, B. Investigation of Anti-Cancer and Migrastatic Properties of Novel Curcumin Derivatives on Breast and Ovarian Cancer Cell Lines. BMC Complement. Altern. Med. 2019, 19, 273. [Google Scholar] [CrossRef]
- Wong, R.S.Y. Apoptosis in Cancer: From Pathogenesis to Treatment. J. Exp. Clin. Cancer Res. 2011, 30, 87. [Google Scholar] [CrossRef] [Green Version]
- Green, D.R.; Bissonnette, R.P.; Cotter, T.G. Apoptosis and Cancer. Important Adv. Oncol. 1994, 1, 37–52. [Google Scholar] [CrossRef]
- Los, M.; Mozoluk, M.; Ferrari, D.; Stepczynska, A.; Stroh, C.; Renz, A.; Herceg, Z.; Wang, Z.-Q.; Schulze-Osthoff, K. Activation and Caspase-Mediated Inhibition of PARP: A Molecular Switch between Fibroblast Necrosis and Apoptosis in Death Receptor Signaling. Mol. Biol. Cell 2002, 13, 978–988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czabotar, P.E.; Lessene, G.; Strasser, A.; Adams, J.M. Control of Apoptosis by the BCL-2 Protein Family: Implications for Physiology and Therapy. Nat. Rev. Mol. Cell Biol. 2013, 15, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Kuwana, T.; Newmeyer, D.D. Bcl-2-Family Proteins and the Role of Mitochondria in Apoptosis. Curr. Opin. Cell Biol. 2003, 15, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Haupt, A.; Joberty, G.; Bantscheff, M.; Fröhlich, H.; Stehr, H.; Schweiger, M.R.; Fischer, A.; Kerick, M.; Boerno, S.T.; Dahl, A.; et al. Hsp90 Inhibition Differentially Destabilises MAP Kinase and TGF-Beta Signalling Components in Cancer Cells Revealed by Kinase-Targeted Chemoproteomics. BMC Cancer 2012, 12, 38. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, S.; Schnellmann, R.G. A Death-Promoting Role for Extracellular Signal-Regulated Kinase. J. Pharmacol. Exp. Ther. 2006, 319, 991–997. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.Y.; Doan, N.D.; DiChiara, A.S.; Papa, L.J., III; Cheah, J.H.; Soule, C.K.; Watson, N.; Hulleman, J.D.; Shoulders, M.D. High-Throughput Assay for Collagen Secretion Suggests an Unanticipated Role for Hsp90 in Collagen Production. Biochemistry 2018, 57, 2814. [Google Scholar] [CrossRef]
- Evan, G.I.; Vousden, K.H. Proliferation, Cell Cycle and Apoptosis in Cancer. Nature 2001, 411, 342–348. [Google Scholar] [CrossRef]
- Nirgude, S.; Desai, S.; Mahadeva, R.; Ravindran, F.; Choudhary, B. ST08 Altered NF-κB Pathway in Breast Cancer Cells In Vitro as Revealed by m S.; Choudha iRNA-mRNA Analysis and Enhanced the Effect of Cisplatin on Tumour Reduction in EAC Mouse Model. Front. Oncol. 2022, 12, 835027. [Google Scholar] [CrossRef] [PubMed]
- Feitelson, M.A.; Arzumanyan, A.; Kulathinal, R.J.; Blain, S.W.; Holcombe, R.F.; Mahajna, J.; Marino, M.; Martinez-Chantar, M.L.; Nawroth, R.; Sanchez-Garcia, I.; et al. Sustained Proliferation in Cancer: Mechanisms and Novel Therapeutic Targets. Semin. Cancer Biol. 2015, 35, S25–S54. [Google Scholar] [CrossRef] [PubMed]
- Schopf, F.H.; Biebl, M.M.; Buchner, J. The HSP90 Chaperone Machinery. Nat. Rev. Mol. Cell Biol. 2017, 18, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, K.S.; Kim, G.P.; Foster, N.R.; Wang-Gillam, A.; Erlichman, C.; McWilliams, R.R. Phase II Trial of Gemcitabine and Tanespimycin (17AAG) in Metastatic Pancreatic Cancer: A Mayo Clinic Phase II Consortium Study. Investig. New Drugs 2015, 33, 963–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, D.L.; Acquaviva, J.; Sequeira, M.; Jimenez, J.P.; Zhang, C.; Sang, J.; Bates, R.C.; Proia, D.A. The HSP90 Inhibitor Ganetespib Potentiates the Antitumor Activity of EGFR Tyrosine Kinase Inhibition in Mutant and Wild-Type Non-Small Cell Lung Cancer. Target. Oncol. 2015, 10, 235–245. [Google Scholar] [CrossRef] [Green Version]
- Jhaveri, K.; Chandarlapaty, S.; Lake, D.; Gilewski, T.; Robson, M.; Goldfarb, S.; Drullinsky, P.; Sugarman, S.; Wasserheit-Leiblich, C.; Fasano, J.; et al. A Phase II Open-Label Study of Ganetespib, a Novel Heat Shock Protein 90 Inhibitor for Patients with Metastatic Breast Cancer. Clin. Breast Cancer 2014, 14, 154–160. [Google Scholar] [CrossRef]
- Koelsch, C.F. Bromination of 3-Acetocoumarin. J. Am. Chem. Soc. 1950, 72, 2993–2995. [Google Scholar] [CrossRef]
- Nirgude, S.; Desai, S.; Choudhary, B. Curcumin Alters Distinct Molecular Pathways in Breast Cancer Subtypes Revealed by Integrated miRNA/mRNA Expression Analysis. Cancer Rep. 2022, 5, e1596. [Google Scholar] [CrossRef]
- Junqueira, L.C.U.; Bignolas, G.; Brentani, R.R. Picrosirius Staining plus Polarization Microscopy, a Specific Method for Collagen Detection in Tissue Sections. Histochem. J. 1979, 11, 447–455. [Google Scholar] [CrossRef]
- Puchtler, H.; Waldrop, F.S.; Valentine, L.S. Polarization Microscopic Studies of Connective Tissue Stained with Picro-Sirius Red FBA. Beiträge Pathol. 1973, 150, 174–187. [Google Scholar] [CrossRef]
- Koroth, J.; Mahadeva, R.; Ravindran, F.; Parashar, T.R.; Teja, V.; Karki, S.S.; Choudhary, B. Curcumin Derivative 1, 2-Bis [(3E, 5E)-3,5-Bis [(2-Chlorophenyl) Methylene]-4-Oxo-1-Piperidyl] Ethane-1,2-Dione (ST03) Induces Mitochondria Mediated Apoptosis in Ovarian Cancer Cells and Inhibits Tumor Progression in EAC Mouse Model. Transl. Oncol. 2021, 15, 101280. [Google Scholar] [CrossRef] [PubMed]













| Cell Line | IC50 (µM) |
|---|---|
| Molt4 | 0.72 |
| Nalm6 | 0.847 |
| CEM | 0.889 |
| Reh | 1.06 |
| K562 | 1.26 |
| CTD | NTD | |
|---|---|---|
| Initial denaturation | 95 °C, 5 min | 95 °C, 5 min |
| Denaturation | 95 °C, 30 s | 95 °C, 30 s |
| Annealing temperature | 50 °C, 35 s | 56.1 °C, 35 s |
| Extension | 72 °C, 40 s | 72 °C, 40 s |
| Final extension | 72 °C, 5 min | 72 °C, 5 min |
| Total no. of cycles | 34 | 34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nirgude, S.; Shahana M. V.; Ravindran, F.; Kumar, S.; Sharma, S.; Mahadeva, R.; Mhatre, A.; Karki, S.S.; Choudhary, B. A Coumarin–Imidazothiadiazole Derivative, SP11 Abrogates Tumor Growth by Targeting HSP90 and Its Client Proteins. Molecules 2023, 28, 5226. https://doi.org/10.3390/molecules28135226
Nirgude S, Shahana M. V., Ravindran F, Kumar S, Sharma S, Mahadeva R, Mhatre A, Karki SS, Choudhary B. A Coumarin–Imidazothiadiazole Derivative, SP11 Abrogates Tumor Growth by Targeting HSP90 and Its Client Proteins. Molecules. 2023; 28(13):5226. https://doi.org/10.3390/molecules28135226
Chicago/Turabian StyleNirgude, Snehal, Shahana M. V., Febina Ravindran, Sujeet Kumar, Shivangi Sharma, Raghunandan Mahadeva, Anisha Mhatre, Subhas S. Karki, and Bibha Choudhary. 2023. "A Coumarin–Imidazothiadiazole Derivative, SP11 Abrogates Tumor Growth by Targeting HSP90 and Its Client Proteins" Molecules 28, no. 13: 5226. https://doi.org/10.3390/molecules28135226
APA StyleNirgude, S., Shahana M. V., Ravindran, F., Kumar, S., Sharma, S., Mahadeva, R., Mhatre, A., Karki, S. S., & Choudhary, B. (2023). A Coumarin–Imidazothiadiazole Derivative, SP11 Abrogates Tumor Growth by Targeting HSP90 and Its Client Proteins. Molecules, 28(13), 5226. https://doi.org/10.3390/molecules28135226