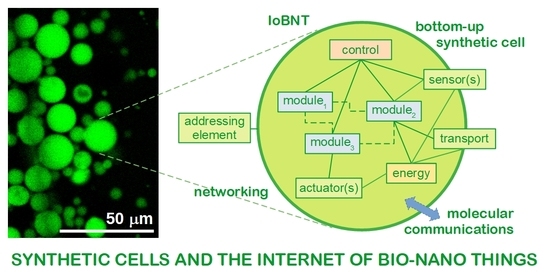
A Role for Bottom-Up Synthetic Cells in the Internet of Bio-Nano Things?
Abstract
:1. Introduction

2. Molecular Communications: History, Perspectives, and Challenges of a Frontier Engineering Approach
3. Internet of Bio-Nano Things (IoBNT)
4. Synthetic Biology and Synthetic Cells
5. Bottom-Up SCs: Principles, Construction, and Features
| Module | Specifications/Possible Realization | Notes |
|---|---|---|
| Sensor | Receptors on the surface and in the lumen | The sensor array selectively (and explicitly) determines which factors the SC will respond to. However, because isolating molecular interactions is not possible, in principle, any SC element can be de facto influenced by third-party molecules. |
| Actuator | Any element or module that can modify the SC dynamics, either to assure SC maintenance, self-distinction, and existence or to achieve a useful programmed goal | It must be activated upon computation. |
| Control, Computation | Switch-like mechanisms, triggers, activation, transistor-like elements [147], chemical neural networks | This should not necessarily be a genetic mechanism; protein functions are often regulated allosterically, and cooperative mechanisms originate a sigmoidal response, signal integration, input classification, recurrence, and self-stabilization. |
| Communication | Actually a combination of sensors, controllers, actuators; explicitly, a module that imitates biological communication mechanisms. More generally, any physicochemical mechanism interpreted as an information transfer device | Examples: covering short-, medium-, and long-range distances (e.g., Ca, molecular motors on filaments, diffusion-driven signaling). |
| Power/Energy | Substrate-level phosphorylation, photo-phosphorylation | The ultimate source of energy must be able to enter the SC, i.e., to be recruited by and to participate in the chemical organization/network. |
6. Engagement of Bottom-Up SCs and Alike Systems in Molecular Communication
6.1. MC Implications of Communicating SCs: Evaluating Syntactic and Semantic Information
6.2. Toward Minimal Cognition
“[a] cognitive structure […] selects, and retroactively alters, the stimuli to which it is sensitive. By this combination of choice and feed-back, the organic structure determines (in a way moulds) its own specific environment; and the environment in turn brings the cognitive organization to its full development. The system and the environment make one another: cognition according to Maturana and Varela is a process of co-emergence”.(italics in the original text) [79]
7. A Role for Bottom-Up Synthetic Cells in the Internet of Bio-Nano Things?
- The constantly improving SC technology and, more specifically, bottom-up approaches. Although in the original IoBNT literature [1], reference was mainly made to “engineered cells” (intended as ad hoc engineered living cells, which are systems obtained by the the top-down SC branch; see Figure 1b and Section 4), our particular interest here lies in the bottom-up approach, recognizing that the corresponding functionalities are certainly inferior to those of the engineered cells. To this aim, SCs can just be seen as rather simple machines, without the need to display the peculiar life-like behaviors or especially the complexity of living cells. Technical advancements from the biochemical/molecular biology perspectives are thus fundamental to reaching these goals. We also suggest that recalling theoretical considerations based on the first and second cybernetics [126,178,179,180] and artificial/biological mirroring [81] (information processing, communication, homeostasis, feedback, autonomy, etc.) can powerfully boost SB approaches: it is possible to learn from explored topics, issues, and solutions in cybernetics and apply them to SB artifacts.
- An accompanying MC theory to model, guide, and drive the above-mentioned SC developments in order to fulfill expectations of the IoBNT scenario. MC is the framework for constructing the IoBNT, whereby one needs to develop strategies that mimic what happens in biological systems and in the (already progressed) IoT.
- Their fruitful combination in a novel, integrated, synergic tool. Elements of the IoBNT can be bio-nanosensors, engineered cells, and bottom-up SCs. The combination of these elements relies on the possibility of interfacing natural or synthetic biodevices with electronic devices, as well as natural biodevices with synthetic biodevices.
8. IoBNT through Neuromorphic Engineering in Wetware
9. Social Robotics in the Chemical Domain: Epistemological Considerations of the Synthetic and Hybrid Ecologies Emerging from Cross-Fertilization between SC and IoBNT
10. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ICTs | Information and communication technologies |
| MC | Molecular communications |
| SB | Synthetic Biology |
| SC | Synthetic Cell |
| IoT | Internet of Things |
| IoBNT | Internet of Bio-Nano Things |
References
- Akyildiz, I.F.; Pierobon, M.; Balasubramaniam, S.; Koucheryavy, Y. The internet of bio-nano things. IEEE Commun. Mag. 2015, 53, 32–40. [Google Scholar] [CrossRef]
- Kusku, M.; Unluturk, B.D. Internet of Bio-Nano Things: A Review of Applications, Enabling Technologies and Key Challenges. ITU J. Future Evol. Technol. 2021, 2, 1–24. [Google Scholar]
- Zafar, S.; Nazir, M.; Bakhshi, T.; Khattak, H.A.; Khan, S.; Bilal, M.; Choo, K.K.R.; Kwak, K.S.; Sabah, A. A systematic review of bio-cyber interface technologies and security issues for internet of bio-nano things. IEEE Access 2021, 9, 93529–93566. [Google Scholar] [CrossRef]
- Nakano, T.; Moore, M.; Enomoto, A.; Suda, T. Molecular Communication Technology as a Biological ICT. In Biological Functions for Information and Communication Technologies; Sawai, H., Ed.; Number 320 in Studies in Computational Intelligence; Springer: Berlin/Heidelberg, Germany, 2011; pp. 49–86. [Google Scholar]
- Nakano, T.; Eckford, A.W.; Haraguchi, T. Molecular Communication; Cambridge University Press: Cambridge, UK, 2013. [Google Scholar]
- Nakano, T. Molecular Communication: A 10 Year Retrospective. IEEE Trans. Mol. Biol. Multi-Scale Commun. 2017, 3, 71–78. [Google Scholar] [CrossRef]
- Akan, O.B.; Ramezani, H.; Khan, T.; Abbasi, N.A.; Kuscu, M. Fundamentals of molecular information and communication science. Proc. IEEE 2016, 105, 306–318. [Google Scholar] [CrossRef]
- Egan, M.; Kuscu, M.; Barros, M.T.; Booth, M.; Llopis-Lorente, A.; Magarini, M.; Martins, D.P.; Schäfer, M.; Stano, P. Toward Interdisciplinary Synergies in Molecular Communications: Perspectives from Synthetic Biology, Nanotechnology, Communications Engineering and Philosophy of Science. Life 2023, 13, 208. [Google Scholar] [CrossRef]
- Stano, P.; Mavelli, F. Protocells Models in Origin of Life and Synthetic Biology. Life 2015, 5, 1700–1702. [Google Scholar] [CrossRef] [Green Version]
- Lentini, R.; Martín, N.Y.; Forlin, M.; Belmonte, L.; Fontana, J.; Cornella, M.; Martini, L.; Tamburini, S.; Bentley, W.E.; Jousson, O.; et al. Two-Way Chemical Communication between Artificial and Natural Cells. ACS Cent. Sci. 2017, 3, 117–123. [Google Scholar] [CrossRef]
- Suda, T.; Moore, M.; Nakano, T.; Egashire, R.; Enomoto, A. Exploratory research on molecular communication between nanomachines. In Proceedings of the Genetic and Evolutionary Computation Conference (GECCO) 2005, Washington DC, USA, 25–29 June 2005; Beyer, H.G., Ed.; ACM Press: New York, NY, USA, 2005; pp. 1–5. [Google Scholar]
- Hiyama, S.; Moritani, Y.; Suda, T.; Egashira, R.; Enamoto, A.; Moore, M.; Nakano, T. Molecular communications. In Proceedings of the 2005 NSTI Nanotechnology Conference and Trade Show, Anaheim, CA, USA, 8–12 May 2005; TechConnect Briefs. Volume 3, pp. 391–394. [Google Scholar]
- Cronin, L.; Krasnogor, N.; Davis, B.G.; Alexander, C.; Robertson, N.; Steinke, J.H.G.; Schroeder, S.L.M.; Khlobystov, A.N.; Cooper, G.; Gardner, P.M.; et al. The imitation game–a computational chemical approach to recognizing life. Nat. Biotechnol. 2006, 24, 1203–1206. [Google Scholar] [CrossRef]
- Damiano, L.; Stano, P. On the “Life-Likeness” of Synthetic Cells. Front. Bioeng. Biotechnol. 2020, 8, 953. [Google Scholar] [CrossRef]
- Leduc, P.R.; Wong, M.S.; Ferreira, P.M.; Groff, R.E.; Haslinger, K.; Koonce, M.P.; Lee, W.Y.; Love, J.C.; McCammon, J.A.; Monteiro-Riviere, N.A.; et al. Towards an in vivo biologically inspired nanofactory. Nat. Nanotechnol. 2007, 2, 3–7. [Google Scholar] [CrossRef]
- Stano, P.; Rampioni, G.; Carrara, P.; Damiano, L.; Leoni, L.; Luisi, P.L. Semi-synthetic minimal cells as a tool for biochemical ICT. BioSystems 2012, 109, 24–34. [Google Scholar] [CrossRef]
- Pierobon, M.; Akyildiz, I.F. Diffusion-Based Noise Analysis for Molecular Communication in Nanonetworks. IEEE Trans. Signal Process. 2011, 59, 2532–2547. [Google Scholar] [CrossRef]
- Kuran, M.; Yilmaz, H.B.; Demirkol, I.; Farsad, N.; Goldsmith, A. A Survey on Modulation Techniques in Molecular Communication via Diffusion. IEEE Commun. Surv. Tutor. 2021, 23, 7–28. [Google Scholar] [CrossRef]
- Jamali, V.; Ahmadzadeh, A.; Wicke, W.; Noel, A.; Schober, R. Channel Modeling for Diffusive Molecular Communication—A Tutorial Review. Proc. IEEE 2019, 107, 1256–1301. [Google Scholar] [CrossRef] [Green Version]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef] [Green Version]
- Farsad, N.; Yilmaz, H.B.; Eckford, A.; Chae, C.B.; Guo, W. A Comprehensive Survey of Recent Advancements in Molecular Communication. IEEE Commun. Surv. Tutor. 2016, 18, 1887–1919. [Google Scholar] [CrossRef] [Green Version]
- Jetka, T.; Nienałtowski, K.; Winarski, T.; Błoński, S.; Komorowski, M. Information-theoretic analysis of multivariate single-cell signaling responses. PLoS Comput. Biol. 2019, 15, e1007132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farsad, N.; Chuang, W.; Goldsmith, A.; Komninakis, C.; Médard, M.; Rose, C.; Vandenberghe, L.; Wesel, E.E.; Wesel, R.D. Capacities and Optimal Input Distributions for Particle-Intensity Channels. IEEE Trans. Mol. Biol. Multi-Scale Commun. 2020, 6, 220–232. [Google Scholar] [CrossRef]
- Ratti, F.; Vakilipoor, F.; Awan, H.; Magarini, M. Bounds on the Constrained Capacity for the Diffusive Poisson Molecular Channel With Memory. IEEE Trans. Mol. Biol. Multi-Scale Commun. 2021, 7, 100–105. [Google Scholar] [CrossRef]
- Johnson, H.A.; Knudsen, K.D. Renal Efficiency and Information Theory. Nature 1965, 206, 930–931. [Google Scholar] [CrossRef]
- Amos, M.; Dittrich, P.; McCaskill, J.; Rasmussen, S. Biological and Chemical Information Technologies. Procedia Comput. Sci. 2011, 7, 56–60. [Google Scholar] [CrossRef]
- Rampioni, G.; Mavelli, F.; Damiano, L.; D’Angelo, F.; Messina, M.; Leoni, L.; Stano, P. A synthetic biology approach to bio-chem-ICT: First moves towards chemical communication between synthetic and natural cells. Nat. Comput. 2014, 13, 333–349. [Google Scholar] [CrossRef]
- Kulakowski, P.; Turbic, K.; Correia, L.M. From nano-communications to body area networks: A perspective on truly personal communications. IEEE Access 2020, 8, 159839–159853. [Google Scholar] [CrossRef]
- Braccini, M.; Collison, E.; Roli, A.; Fellermann, H.; Stano, P. Recurrent Neural Networks in Synthetic Cells: A Route to Autonomous Molecular Agents? Front. Bioeng. Biotechnol. 2023, 11, 1210334. [Google Scholar] [CrossRef] [PubMed]
- Guidoni, A. Verso il Robot Sapiens, 1st ed.; Collana Scienza, Edizioni Controluce; Monte Compatri: Rome, Italy, 2018. [Google Scholar]
- Magarini, M.; Stano, P. Synthetic Cells Engaged in Molecular Communication: An Opportunity for Modelling Shannon- and Semantic-Information in the Chemical Domain. Front. Commun. Netw. 2021, 2, 48. [Google Scholar] [CrossRef]
- Stano, P. Exploring Information and Communication Theories for Synthetic Cell Research. Front. Bioeng. Biotechnol. 2022, 10, 927156. [Google Scholar] [CrossRef]
- Leduc, S. La Biologie Synthétique, 1st ed.; Etudes de Biophysique, A. Poinat: Paris, France, 1912. [Google Scholar]
- Szybalski, W. In Vivo and In Vitro Initiation of Transcription. In Control of Gene Expression; Kohn, A., Shatkay, A., Eds.; Advances in Experimental Medicine and Biology; Springer US: Boston, MA, USA, 1974; Volume 44, pp. 23–24. [Google Scholar] [CrossRef]
- Aloni, Y.; Aviv, H.; Bautz, E.K.F.; Bizunski, M.; Conconi, F.; Daniel, V.; Elson, D.; Herzberg, M.; Inouye, H.; Kaye, A.M.; et al. Panel Discussion. In Control of Gene Expression; Kohn, A., Shatkay, A., Eds.; Advances in Experimental Medicine and Biology; Springer US: Boston, MA, USA, 1974; Volume 44, pp. 403–420. [Google Scholar] [CrossRef]
- Luisi, P.L. Chemical aspects of synthetic biology. Chem. Biodivers. 2007, 4, 603–621. [Google Scholar] [CrossRef]
- Chiarabelli, C.; Stano, P.; Luisi, P.L. Chemical synthetic biology: A mini-review. Front. Microbiol. 2013, 4, 285. [Google Scholar] [CrossRef] [Green Version]
- Perez, J.G.; Stark, J.C.; Jewett, M.C. Cell-Free Synthetic Biology: Engineering Beyond the Cell. Cold Spring Harb. Perspect. Biol. 2016, 8, a023853. [Google Scholar] [CrossRef] [Green Version]
- Tinafar, A.; Jaenes, K.; Pardee, K. Synthetic Biology Goes Cell-Free. BMC Biol. 2019, 17, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Kwon, Y.C.; Lu, Y.; Moore, S.J. Editorial: Cell-Free Synthetic Biology. Front. Bioeng. Biotechnol. 2021, 9, 799122. [Google Scholar] [CrossRef]
- Bedau, M.A.; Parke, E.C.; Tangen, U.; Hantsche-Tangen, B. Social and ethical checkpoints for bottom-up synthetic biology, or protocells. Syst. Synth. Biol. 2009, 3, 65–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luisi, P.L. The Synthetic Approach in Biology: Epistemological Notes for Synthetic Biology. In Chemical Synthetic Biology; Luisi, P.L., Chiarabelli, C., Eds.; Wiley: Chichester, NY, USA, 2011; pp. 343–362. [Google Scholar]
- Deplazes, A.; Huppenbauer, M. Synthetic organisms and living machines: Positioning the products of synthetic biology at the borderline between living and non-living matter. Syst. Synth. Biol. 2009, 3, 55–63. [Google Scholar] [CrossRef] [Green Version]
- Deplazes-Zemp, A. Artificial Cell Research as a Field that Connects Chemical, Biological and Philosophical Questions. Chimia 2016, 70, 443–448. [Google Scholar] [CrossRef] [Green Version]
- Damiano, L.; Stano, P. Explorative Synthetic Biology in AI. Criteria of relevance and a taxonomy for synthetic models of living and cognitive processes. Artif. Life, 2023; in press. [Google Scholar]
- Stano, P.; Damiano, L. Synthetic cell research: Is technical progress leaving theoretical and epistemological investigations one step behind? Front. Robot. AI 2023, 10, 1143196. [Google Scholar] [CrossRef] [PubMed]
- Luisi, P.L. The Emergence of Life: From Chemical Origins to Synthetic Biology, 1st ed.; Cambridge University Press: Cambridge, UK, 2006. [Google Scholar]
- Noireaux, V.; Bar-Ziv, R.; Libchaber, A. Principles of cell-free genetic circuit assembly. Proc. Natl. Acad. Sci. USA 2003, 100, 12672–12677. [Google Scholar] [CrossRef]
- Shin, J.; Noireaux, V. An E. coli Cell-Free Expression Toolbox: Application to Synthetic Gene Circuits and Artificial Cells. ACS Synth. Biol. 2012, 1, 29–41. [Google Scholar] [CrossRef]
- Sleator, R.D. The story of Mycoplasma mycoides JCVI-syn1.0: The forty million dollar microbe. Bioeng. Bugs 2010, 1, 229–230. [Google Scholar] [CrossRef] [Green Version]
- Gibson, D.G.; Glass, J.I.; Lartigue, C.; Noskov, V.N.; Chuang, R.Y.; Algire, M.A.; Benders, G.A.; Montague, M.G.; Ma, L.; Moodie, M.M.; et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 2010, 329, 52–56. [Google Scholar] [CrossRef] [Green Version]
- Glass, J.I. Synthetic genomics and the construction of a synthetic bacterial cell. Perspect. Biol. Med. 2012, 55, 473–489. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, C.A.; Chuang, R.Y.; Noskov, V.N.; Assad-Garcia, N.; Deerinck, T.J.; Ellisman, M.H.; Gill, J.; Kannan, K.; Karas, B.J.; Ma, L.; et al. Design and synthesis of a minimal bacterial genome. Science 2016, 351, aad6253. [Google Scholar] [CrossRef] [Green Version]
- Pelletier, J.F.; Sun, L.; Wise, K.S.; Assad-Garcia, N.; Karas, B.J.; Deerinck, T.J.; Ellisman, M.H.; Mershin, A.; Gershenfeld, N.; Chuang, R.Y.; et al. Genetic requirements for cell division in a genomically minimal cell. Cell 2021, 184, 2430–2440.e16. [Google Scholar] [CrossRef] [PubMed]
- Thornburg, Z.R.; Bianchi, D.M.; Brier, T.A.; Gilbert, B.R.; Earnest, T.M.; Melo, M.C.R.; Safronova, N.; Sáenz, J.P.; Cook, A.T.; Wise, K.S.; et al. Fundamental behaviors emerge from simulations of a living minimal cell. Cell 2022, 185, 345–360.e28. [Google Scholar] [CrossRef]
- Endy, D. Foundations for engineering biology. Nature 2005, 438, 449–453. [Google Scholar] [CrossRef]
- de Lorenzo, V.; Danchin, A. Synthetic biology: Discovering new worlds and new words. EMBO Rep. 2008, 9, 822–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Hu, S.; Chen, X. Artificial cells: From basic science to applications. Mater. Today 2016, 19, 516–532. [Google Scholar] [CrossRef]
- Buddingh’, B.C.; van Hest, J.C.M. Artificial Cells: Synthetic Compartments with Life-like Functionality and Adaptivity. Acc. Chem. Res. 2017, 50, 769–777. [Google Scholar] [CrossRef] [Green Version]
- Salehi-Reyhani, A.; Ces, O.; Elani, Y. Artificial cell mimics as simplified models for the study of cell biology. Exp. Biol. Med. 2017, 242, 1309–1317. [Google Scholar] [CrossRef] [Green Version]
- Schwille, P.; Spatz, J.; Landfester, K.; Bodenschatz, E.; Herminghaus, S.; Sourjik, V.; Erb, T.J.; Bastiaens, P.; Lipowsky, R.; Hyman, A.; et al. MaxSynBio: Avenues Towards Creating Cells from the Bottom Up. Angew. Chem. Int. Ed. Engl. 2018, 57, 13382–13392. [Google Scholar] [CrossRef]
- Stano, P. Gene Expression Inside Liposomes: From Early Studies to Current Protocols. Chemistry 2019, 25, 7798–7814. [Google Scholar] [CrossRef]
- Stano, P. Is Research on “Synthetic Cells” Moving to the Next Level? Life 2019, 9, 3. [Google Scholar] [CrossRef] [Green Version]
- Abil, Z.; Danelon, C. Roadmap to Building a Cell: An Evolutionary Approach. Front. Bioeng. Biotechnol. 2020, 8, 927. [Google Scholar] [CrossRef] [PubMed]
- Gaut, N.J.; Adamala, K.P. Reconstituting Natural Cell Elements in Synthetic Cells. Adv. Biol. 2021, 5, e2000188. [Google Scholar] [CrossRef]
- Mukwaya, V.; Mann, S.; Dou, H. Chemical communication at the synthetic cell/living cell interface. Commun. Chem. 2021, 4, 161. [Google Scholar] [CrossRef]
- Staufer, O.; De Lora, J.A.; Bailoni, E.; Bazrafshan, A.; Benk, A.S.; Jahnke, K.; Manzer, Z.A.; Otrin, L.; Díez Pérez, T.; Sharon, J.; et al. Building a community to engineer synthetic cells and organelles from the bottom-up. Elife 2021, 10, e73556. [Google Scholar] [CrossRef] [PubMed]
- Herianto, S.; Chien, P.J.; Ho, J.a.A.; Tu, H.L. Liposome-based artificial cells: From gene expression to reconstitution of cellular functions and phenotypes. Biomater. Adv. 2022, 142, 213156. [Google Scholar] [CrossRef] [PubMed]
- Hirschi, S.; Ward, T.R.; Meier, W.P.; Müller, D.J.; Fotiadis, D. Synthetic Biology: Bottom-Up Assembly of Molecular Systems. Chem. Rev. 2022, 122, 16294–16328. [Google Scholar] [CrossRef]
- Ghosh, B. Artificial cell design: Reconstructing biology for life science applications. Emerg. Top. Life Sci. 2022, 6, 619–627. [Google Scholar] [CrossRef]
- Guindani, C.; da Silva, L.C.; Cao, S.; Ivanov, T.; Landfester, K. Synthetic Cells: From Simple Bio-Inspired Modules to Sophisticated Integrated Systems. Angew. Chem. Int. Ed. Engl. 2022, 61, e202110855. [Google Scholar] [CrossRef]
- Jiang, W.; Wu, Z.; Gao, Z.; Wan, M.; Zhou, M.; Mao, C.; Shen, J. Artificial Cells: Past, Present and Future. ACS Nano 2022, 16, 15705–15733. [Google Scholar] [CrossRef]
- Nicholson, D.J. Organisms≠Machines. Stud. Hist. Philos. Sci. Part C Stud. Hist. Philos. Biol. Biomed. Sci. 2013, 44, 669–678. [Google Scholar] [CrossRef]
- Stano, P. A four-track perspective for bottom-up synthetic cells. Front. Bioeng. Biotechnol. 2022, 10, 1029446. [Google Scholar] [CrossRef]
- Maturana, H.R.; Varela, F.J. Autopoiesis and Cognition: The Realization of the Living, 1st ed.; D. Reidel Publishing Company: Dordrecht, The Netherlands, 1980. [Google Scholar]
- Luisi, P.L. Autopoiesis: A review and a reappraisal. Naturwissenschaften 2003, 90, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Luisi, P.L.; Ferri, F.; Stano, P. Approaches to semi-synthetic minimal cells: A review. Naturwissenschaften 2006, 93, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Stano, P. From Chemical Autopoiesis to Synthetic Biology. L’Actualité Chim. 2020, 455, 31–40. [Google Scholar]
- Bitbol, M.; Luisi, P.L. Autopoiesis with or without cognition: Defining life at its edge. J. R. Soc. Interface 2004, 1, 99–107. [Google Scholar] [CrossRef]
- Bourgine, P.; Stewart, J. Autopoiesis and cognition. Artif. Life 2004, 10, 327–345. [Google Scholar] [CrossRef] [PubMed]
- Cordeschi, R. The Discovery of the Artificial: Behavior, Mind and Machines Before and Beyond Cybernetics; Studies in Cognitive Systems; Springer: Amsterdam, The Netherlands, 2002. [Google Scholar]
- Damiano, L.; Hiolle, A.; Cañamero, L. Grounding Synthetic Knowledge. In Advances in Artificial Life, ECAL 2011; Lenaerts, T., Giacobini, M., Bersini, H., Bourgine, P., Dorigo, M., Doursat, R., Eds.; MIT Press: Cambrigde, MA, USA, 2011; pp. 200–207. [Google Scholar]
- Damiano, L.; Stano, P. Synthetic Biology and Artificial Intelligence. Grounding a cross-disciplinary approach to the synthetic exploration of (embodied) cognition. Complex Syst. 2018, 27, 199–228. [Google Scholar] [CrossRef]
- Bich, L.; Frick, R. Synthetic Modelling of Biological Communication: A Theoretical and Operational Framework for the Investigation of Minimal Life and Cognition. Complex Syst. 2018, 27, 267–287. [Google Scholar] [CrossRef]
- Frick, R.; Bich, L.; Moreno, A. An Organisational Approach to Biological Communication. Acta Biotheor. 2019, 67, 103–128. [Google Scholar] [CrossRef] [PubMed]
- Damiano, L.; Stano, P. A Wetware Embodied AI? Towards an Autopoietic Organizational Approach Grounded in Synthetic Biology. Front. Bioeng. Biotechnol. 2021, 9, 873. [Google Scholar] [CrossRef]
- Oberholzer, T.; Wick, R.; Luisi, P.L.; Biebricher, C.K. Enzymatic RNA replication in self-reproducing vesicles: An approach to a minimal cell. Biochem. Biophys. Res. Commun. 1995, 207, 250–257. [Google Scholar] [CrossRef]
- Szostak, J.W.; Bartel, D.P.; Luisi, P.L. Synthesizing life. Nature 2001, 409, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.A.; Salehi-Ashtiani, K.; Szostak, J.W. RNA catalysis in model protocell vesicles. J. Am. Chem. Soc. 2005, 127, 13213–13219. [Google Scholar] [CrossRef] [Green Version]
- Mansy, S.S.; Szostak, J.W. Reconstructing the emergence of cellular life through the synthesis of model protocells. Cold Spring Harb. Symp. Quant. Biol. 2009, 74, 47–54. [Google Scholar] [CrossRef]
- Cho, E.; Lu, Y. Compartmentalizing Cell-Free Systems: Toward Creating Life-Like Artificial Cells and Beyond. ACS Synth. Biol. 2020, 9, 2881–2901. [Google Scholar] [CrossRef]
- Olivi, L.; Berger, M.; Creyghton, R.N.P.; De Franceschi, N.; Dekker, C.; Mulder, B.M.; Claassens, N.J.; Ten Wolde, P.R.; van der Oost, J. Towards a synthetic cell cycle. Nat. Commun. 2021, 12, 4531. [Google Scholar] [CrossRef] [PubMed]
- Gentili, P.L.; Stano, P. Monitoring the advancements in the technology of artificial cells by determining their complexity degree: Hints from complex systems descriptors. Front. Bioeng. Biotechnol. 2023, 11, 1132546. [Google Scholar] [CrossRef]
- Szoka, F.; Papahadjopoulos, D. Comparative properties and methods of preparation of lipid vesicles (liposomes). Annu. Rev. Biophys. Bioeng. 1980, 9, 467–508. [Google Scholar] [CrossRef]
- New, R.R.C. Liposomes: A Practical Approach, 1st ed.; IRL Press at Oxford University Press: Oxford, UK, 1990. [Google Scholar]
- Walde, P. Preparation of Vesicles (Liposomes). In Encyclopedia of Nanoscience and Nanotechnology; Nalwa, H.S., Ed.; American Scientific Publishers: Valencia, CA, USA, 2004; Volume 9, pp. 43–79. [Google Scholar]
- Luisi, P.L.; Walde, P. (Eds.) Giant Vesicles; Wiley: Chichester, UK; New York, NY, USA, 2000. [Google Scholar]
- Walde, P.; Cosentino, K.; Engel, H.; Stano, P. Giant vesicles: Preparations and applications. Chembiochem 2010, 11, 848–865. [Google Scholar] [CrossRef]
- Dimova, R.; Stano, P.; Marques, C.M.; Walde, P. Preparation methods for giant unilamellar vesicles. In The Giant Vesicle Book; Dimova, R., Marques, C.M., Eds.; Taylor & Francis Group: Boca Raton, FL, USA, 2020; pp. 3–20. [Google Scholar]
- Pautot, S.; Frisken, B.J.; Weitz, D.A. Production of unilamellar vesicles using an inverted emulsion. Langmuir 2003, 19, 2870–2879. [Google Scholar] [CrossRef]
- Noireaux, V.; Libchaber, A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl. Acad. Sci. USA 2004, 101, 17669–17674. [Google Scholar] [CrossRef] [PubMed]
- Stano, P. Commentary: Rapid and facile preparation of giant vesicles by the droplet transfer method for artificial cell construction. Front. Bioeng. Biotechnol. 2022, 10, 873854. [Google Scholar] [CrossRef] [PubMed]
- Pereira de Souza, T.; Stano, P.; Luisi, P.L. The minimal size of liposome-based model cells brings about a remarkably enhanced entrapment and protein synthesis. Chembiochem 2009, 10, 1056–1063. [Google Scholar] [CrossRef]
- Stano, P. Minimal cells: Relevance and interplay of physical and biochemical factors. Biotechnol. J. 2011, 6, 850–859. [Google Scholar] [CrossRef]
- Pols, T.; Sikkema, H.R.; Gaastra, B.F.; Frallicciardi, J.; Śmigiel, W.M.; Singh, S.; Poolman, B. A synthetic metabolic network for physicochemical homeostasis. Nat. Commun. 2019, 10, 4239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailoni, E.; Partipilo, M.; Coenradij, J.; Grundel, D.A.J.; Slotboom, D.J.; Poolman, B. Minimal Out-of-Equilibrium Metabolism for Synthetic Cells: A Membrane Perspective. ACS Synth. Biol. 2023, 12, 922–946. [Google Scholar] [CrossRef]
- Shimizu, Y.; Inoue, A.; Tomari, Y.; Suzuki, T.; Yokogawa, T.; Nishikawa, K.; Ueda, T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001, 19, 751–755. [Google Scholar] [CrossRef]
- Kuruma, Y.; Stano, P.; Ueda, T.; Luisi, P.L. A synthetic biology approach to the construction of membrane proteins in semi-synthetic minimal cells. Biochim. Biophys. Acta 2009, 1788, 567–574. [Google Scholar] [CrossRef] [Green Version]
- Gessesse, B.; Nagaike, T.; Nagata, K.; Shimizu, Y.; Ueda, T. G-Protein Coupled Receptor Protein Synthesis on a Lipid Bilayer Using a Reconstituted Cell-Free Protein Synthesis System. Life 2018, 8, 54. [Google Scholar] [CrossRef] [Green Version]
- Amati, A.M.; Graf, S.; Deutschmann, S.; Dolder, N.; von Ballmoos, C. Current problems and future avenues in proteoliposome research. Biochem. Soc. Trans. 2020, 48, 1473–1492. [Google Scholar] [CrossRef] [PubMed]
- Kruyer, N.S.; Sugianto, W.; Tickman, B.I.; Alba Burbano, D.; Noireaux, V.; Carothers, J.M.; Peralta-Yahya, P. Membrane Augmented Cell-Free Systems: A New Frontier in Biotechnology. ACS Synth. Biol. 2021, 10, 670–681. [Google Scholar] [CrossRef] [PubMed]
- Nakai, H.; Isshiki, K.; Hattori, M.; Maehira, H.; Yamaguchi, T.; Masuda, K.; Shimizu, Y.; Watanabe, T.; Hohsaka, T.; Shihoya, W.; et al. Cell-Free Synthesis of Human Endothelin Receptors and Its Application to Ribosome Display. Anal. Chem. 2022, 94, 3831–3839. [Google Scholar] [CrossRef] [PubMed]
- Hellingwerf, K.J.; Postma, P.W.; Tommassen, J.; Westerhoff, H.V. Signal transduction in bacteria: Phospho-neural network(s) in Escherichia coli? FEMS Microbiol. Rev. 1995, 16, 309–321. [Google Scholar] [CrossRef]
- Gentili, P.L.; Stano, P. Chemical Neural Networks Inside Synthetic Cells? A Proposal for Their Realization and Modeling. Front. Bioeng. Biotechnol. 2022, 10, 927110. [Google Scholar] [CrossRef]
- Siegal-Gaskins, D.; Tuza, Z.A.; Kim, J.; Noireaux, V.; Murray, R.M. Gene circuit performance characterization and resource usage in a cell-free “breadboard”. ACS Synth. Biol. 2014, 3, 416–425. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, M.K.; Hayes, C.A.; Chappell, J.; Sun, Z.Z.; Murray, R.M.; Noireaux, V.; Lucks, J.B. Characterizing and prototyping genetic networks with cell-free transcription–translation reactions. Methods 2015, 86, 60–72. [Google Scholar] [CrossRef] [Green Version]
- Singhal, V.; Tuza, Z.A.; Sun, Z.Z.; Murray, R.M. A MATLAB toolbox for modeling genetic circuits in cell-free systems. Synth. Biol. 2021, 6, ysab007. [Google Scholar] [CrossRef]
- Crowther, M.; Wipat, A.; Goñi-Moreno, A. A Network Approach to Genetic Circuit Designs. ACS Synth. Biol. 2022, 11, 3058–3066. [Google Scholar] [CrossRef] [PubMed]
- Sents, Z.; Stoughton, T.E.; Buecherl, L.; Thomas, P.J.; Fontanarrosa, P.; Myers, C.J. SynBioSuite: A Tool for Improving the Workflow for Genetic Design and Modeling. ACS Synth. Biol. 2023, 12, 892–897. [Google Scholar] [CrossRef] [PubMed]
- Mavelli, F.; Marangoni, R.; Stano, P. A Simple Protein Synthesis Model for the PURE System Operation. Bull. Math. Biol. 2015, 77, 1185–1212. [Google Scholar] [CrossRef]
- Mavelli, F.; Stano, P. Experiments on and Numerical Modeling of the Capture and Concentration of Transcription-Translation Machinery inside Vesicles. Artif. Life 2015, 21, 445–463. [Google Scholar] [CrossRef]
- Dupin, A.; Aufinger, L.; Styazhkin, I.; Rothfischer, F.; Kaufmann, B.K.; Schwarz, S.; Galensowske, N.; Clausen-Schaumann, H.; Simmel, F.C. Synthetic cell-based materials extract positional information from morphogen gradients. Sci. Adv. 2022, 8, eabl9228. [Google Scholar] [CrossRef]
- Ruzzante, B.; Del Moro, L.; Magarini, M.; Stano, P. Synthetic Cells Extract Semantic Information from their Environment. IEEE Trans. Mol. Biol. Multi-Scale Commun. 2023, 9, 23–27. [Google Scholar] [CrossRef]
- Kolchinsky, A.; Wolpert, D.H. Semantic information, autonomous agency and non-equilibrium statistical physics. Interface Focus 2018, 8, 20180041. [Google Scholar] [CrossRef]
- MacKay, D.M. Information, Mechanism and Meaning; MIT Press: Cambridge, MA, USA, 1969. [Google Scholar]
- Varela, F.J. Principles of Biological Autonomy; The North-Holland Series in General Systems Research; Elsevier North-Holland, Inc.: New York, NY, USA, 1979. [Google Scholar]
- Ruiz-Mirazo, K.; Moreno, A. Basic autonomy as a fundamental step in the synthesis of life. Artif. Life 2004, 10, 235–259. [Google Scholar] [CrossRef]
- Bertschinger, N.; Olbrich, E.; Ay, N.; Jost, J. Autonomy: An information theoretic perspective. Biosystems 2008, 91, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.; Mossio, M. Biological Autonomy: A Philosophical and Theoretical Enquiry; History, Philosophy and Theory of the Life Sciences (HPTL); Springer: Cham, Switzerland, 2015; Volume 12. [Google Scholar]
- Atlan, H. Self Creation of Meaning. Phys. Scr. 1987, 36, 563–576. [Google Scholar] [CrossRef]
- Stano, P. Sketching How Synthetic Cells Can Function as a Platform to Investigate Chemical AI and Information Theories in the Wetware Domain. In Proceedings of the Fourth International Conference on Communication, Computing and Electronics Systems, Haldia, India, 16–18 March 2023; Lecture Notes in Electrical, Engineering. Bindhu, V., Tavares, J.M.R.S., Vuppalapati, C., Eds.; Springer Nature: Singapore, 2023; pp. 571–584. [Google Scholar] [CrossRef]
- Luisi, P.L.; Varela, F.J. Self-replicating micelles—A chemical version of a minimal autopoietic system. Origins Life Evol. Biosphere 1989, 19, 633–643. [Google Scholar] [CrossRef]
- Schmidli, P.K.; Schurtenberger, P.; Luisi, P.L. Liposome-mediated enzymatic synthesis of phosphatidylcholine as an approach to self-replicating liposomes. J. Am. Chem. Soc. 1991, 113, 8127–8130. [Google Scholar] [CrossRef]
- Hanczyc, M.M. The Early History of Protocells—The search for the recipe of life. In Protocells: Bridging Nonliving and Living Matter; Rasmussen, S., Bedau, M.A., Chen, L., Deamer, D., Krakauer, D.C., Packard, N.H., Stadler, P.F., Eds.; MIT Press: Cambridge, MA, USA, 2009; pp. 3–18. [Google Scholar]
- Stano, P. The birth of liposome-based synthetic biology: A brief account. In Liposomes: Historical, Clinical and Molecular Perspectives; Pearson, B.R., Ed.; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2017; pp. 37–52. [Google Scholar]
- Eto, S.; Matsumura, R.; Shimane, Y.; Fujimi, M.; Berhanu, S.; Kasama, T.; Kuruma, Y. Phospholipid synthesis inside phospholipid membrane vesicles. Commun. Biol. 2022, 5, 1016. [Google Scholar] [CrossRef] [PubMed]
- Ramundo-Orlando, A.; Serafino, A.; Villalobo, A. Gap junction channels reconstituted in two closely apposed lipid bilayers. Arch. Biochem. Biophys. 2005, 436, 128–135. [Google Scholar] [CrossRef] [Green Version]
- Kaneda, M.; Nomura, S.i.M.; Ichinose, S.; Kondo, S.; Nakahama, K.i.; Akiyoshi, K.; Morita, I. Direct formation of proteo-liposomes by in vitro synthesis and cellular cytosolic delivery with connexin-expressing liposomes. Biomaterials 2009, 30, 3971–3977. [Google Scholar] [CrossRef]
- Takada, S.; Yoshinaga, N.; Doi, N.; Fujiwara, K. Controlling the Periodicity of a Reaction–Diffusion Wave in Artificial Cells by a Two-Way Energy Supplier. ACS Nano 2022, 16, 16853–16861. [Google Scholar] [CrossRef]
- Krinsky, N.; Kaduri, M.; Zinger, A.; Shainsky-Roitman, J.; Goldfeder, M.; Benhar, I.; Hershkovitz, D.; Schroeder, A. Synthetic Cells Synthesize Therapeutic Proteins inside Tumors. Adv. Healthc. Mater. 2018, 7, e1701163. [Google Scholar] [CrossRef]
- Ding, Y.; Contreras-Llano, L.E.; Morris, E.; Mao, M.; Tan, C. Minimizing Context Dependency of Gene Networks Using Artificial Cells. ACS Appl. Mater. Interfaces 2018, 10, 30137–30146. [Google Scholar] [CrossRef]
- Diltemiz, S.E.; Tavafoghi, M.; Barros, N.R.d.; Kanada, M.; Heinämäki, J.; Contag, C.; Seidlits, S.K.; Ashammakhi, N. Use of artificial cells as drug carriers. Mater. Chem. Front. 2021, 5, 6672–6692. [Google Scholar] [CrossRef]
- Lee, K.Y.; Park, S.J.; Lee, K.A.; Kim, S.H.; Kim, H.; Meroz, Y.; Mahadevan, L.; Jung, K.H.; Ahn, T.K.; Parker, K.K.; et al. Photosynthetic artificial organelles sustain and control ATP-dependent reactions in a protocellular system. Nat. Biotechnol. 2018, 36, 530–535. [Google Scholar] [CrossRef]
- Berhanu, S.; Ueda, T.; Kuruma, Y. Artificial photosynthetic cell producing energy for protein synthesis. Nat. Commun. 2019, 10, 1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altamura, E.; Albanese, P.; Marotta, R.; Milano, F.; Fiore, M.; Trotta, M.; Stano, P.; Mavelli, F. Chromatophores efficiently promote light-driven ATP synthesis and DNA transcription inside hybrid multicompartment artificial cells. Proc. Natl. Acad. Sci. USA 2021, 118, e2012170118. [Google Scholar] [CrossRef] [PubMed]
- Partipilo, M.; Claassens, N.J.; Slotboom, D.J. A Hitchhiker’s Guide to Supplying Enzymatic Reducing Power into Synthetic Cells. ACS Synth. Biol. 2023, 12, 947–962. [Google Scholar] [CrossRef] [PubMed]
- Bray, D. Wetware. A Computer in Every Living Cells; Yale University Press: New Haven, CT, USA; London, UK, 2009. [Google Scholar]
- Mataric, M.J. The Robotics Primer: Intelligent Robotics and Autonomous Agents; MIT Press: Cambridge, MA, USA, 2007. [Google Scholar]
- Rampioni, G.; D’Angelo, F.; Leoni, L.; Stano, P. Gene-Expressing Liposomes as Synthetic Cells for Molecular Communication Studies. Front. Bioeng. Biotechnol. 2019, 7, 1. [Google Scholar] [CrossRef]
- Lentini, R.; Santero, S.P.; Chizzolini, F.; Cecchi, D.; Fontana, J.; Marchioretto, M.; Del Bianco, C.; Terrell, J.L.; Spencer, A.C.; Martini, L.; et al. Integrating artificial with natural cells to translate chemical messages that direct E. coli behaviour. Nat. Commun. 2014, 5, 4012. [Google Scholar] [CrossRef] [Green Version]
- Adamala, K.P.; Martin-Alarcon, D.A.; Guthrie-Honea, K.R.; Boyden, E.S. Engineering genetic circuit interactions within and between synthetic minimal cells. Nat. Chem. 2017, 9, 431–439. [Google Scholar] [CrossRef] [Green Version]
- Gines, G.; Zadorin, A.S.; Galas, J.C.; Fujii, T.; Estevez-Torres, A.; Rondelez, Y. Microscopic agents programmed by DNA circuits. Nat. Nanotech. 2017, 12, 351–359. [Google Scholar] [CrossRef]
- Rampioni, G.; D’Angelo, F.; Messina, M.; Zennaro, A.; Kuruma, Y.; Tofani, D.; Leoni, L.; Stano, P. Synthetic cells produce a quorum sensing chemical signal perceived by Pseudomonas aeruginosa. Chem. Commun. 2018, 54, 2090–2093. [Google Scholar] [CrossRef]
- Niederholtmeyer, H.; Chaggan, C.; Devaraj, N.K. Communication and quorum sensing in non-living mimics of eukaryotic cells. Nat. Commun. 2018, 9, 5027. [Google Scholar] [CrossRef] [Green Version]
- Dupin, A.; Simmel, F.C. Signalling and differentiation in emulsion-based multi-compartmentalized in vitro gene circuits. Nat. Chem. 2019, 11, 32–39. [Google Scholar] [CrossRef]
- Joesaar, A.; Yang, S.; Bögels, B.; van der Linden, A.; Pieters, P.; Kumar, B.V.V.S.P.; Dalchau, N.; Phillips, A.; Mann, S.; de Greef, T.F.A. DNA-based communication in populations of synthetic protocells. Nat. Nanotechnol. 2019, 14, 369–378. [Google Scholar] [CrossRef]
- Buddingh’, B.C.; Elzinga, J.; van Hest, J.C.M. Intercellular communication between artificial cells by allosteric amplification of a molecular signal. Nat. Commun. 2020, 11, 1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Tian, L.; Ren, Y.; Zhao, Z.; Du, H.; Zhang, Z.; Drinkwater, B.W.; Mann, S.; Han, X. Chemical Information Exchange in Organized Protocells and Natural Cell Assemblies with Controllable Spatial Positions. Small 2020, 16, e1906394. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Pieters, P.A.; Joesaar, A.; Bögels, B.W.A.; Brouwers, R.; Myrgorodska, I.; Mann, S.; de Greef, T.F.A. Light-Activated Signaling in DNA-Encoded Sender-Receiver Architectures. ACS Nano 2020, 14, 15992–16002. [Google Scholar] [CrossRef]
- Chen, C.; Wang, X.; Wang, Y.; Tian, L.; Cao, J. Construction of protocell-based artificial signal transduction pathways. Chem. Commun. 2021, 57, 12754–12763. [Google Scholar] [CrossRef] [PubMed]
- Gispert, I.; Hindley, J.W.; Pilkington, C.P.; Shree, H.; Barter, L.M.C.; Ces, O.; Elani, Y. Stimuli-responsive vesicles as distributed artificial organelles for bacterial activation. Proc. Natl. Acad. Sci. USA 2022, 119, e2206563119. [Google Scholar] [CrossRef]
- Zambrano, A.; Fracasso, G.; Gao, M.; Ugrinic, M.; Wang, D.; Appelhans, D.; deMello, A.; Tang, T.Y.D. Programmable synthetic cell networks regulated by tuneable reaction rates. Nat. Commun. 2022, 13, 3885. [Google Scholar] [CrossRef]
- Llopis-Lorente, A.; Buddingh’, B.C.; Martínez-Máñez, R.; van Hest, J.C.M.; Abdelmohsen, L.K.E. Quorum sensing communication between lipid-based artificial cells. Chem. Commun. 2023, 59, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Ariga, K.; Leong, D.T.; Mori, T. Nanoarchitectonics for hybrid and related materials for bio-oriented applications. Adv. Funct. Mater. 2018, 28, 1702905. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, L.; Lim, K.H.; Gonuguntla, S.; Lim, K.W.; Pranantyo, D.; Yong, W.P.; Yam, W.J.T.; Low, Z.; Teo, W.J.; et al. The Pathway to Intelligence: Using Stimuli-Responsive Materials as Building Blocks for Constructing Smart and Functional Systems. Adv. Mater. 2019, 31, 1804540. [Google Scholar] [CrossRef]
- Giménez, C.; Climent, E.; Aznar, E.; Martínez-Máñez, R.; Sancenón, F.; Marcos, M.D.; Amorós, P.; Rurack, K. Towards chemical communication between gated nanoparticles. Angew. Chem. Int. Ed. 2014, 53, 12629–12633. [Google Scholar] [CrossRef]
- de Luis, B.; Llopis-Lorente, A.; Rincón, P.; Gadea, J.; Sancenón, F.; Aznar, E.; Villalonga, R.; Murguía, J.R.; Martínez-Máñez, R. An interactive model of communication between abiotic nanodevices and microorganisms. Angew. Chem. Int. Ed. 2019, 58, 14986–14990. [Google Scholar] [CrossRef]
- de Luis, B.; Morellá-Aucejo, Á.; Llopis-Lorente, A.; Martínez-Latorre, J.; Sancenón, F.; López, C.; Murguía, J.R.; Martínez-Máñez, R. Nanoprogrammed Cross-Kingdom Communication Between Living Microorganisms. Nano Lett. 2022, 22, 1836–1844. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.M.S. Artificial Cells; Charles C. Thomas: Springfield, IL, USA, 1972. [Google Scholar]
- Logan, R.K. What Is Information?: Why Is It Relativistic and What Is Its Relationship to Materiality, Meaning and Organization. Information 2012, 3, 68–91. [Google Scholar] [CrossRef] [Green Version]
- Hayles, N.K. How We Became Posthuman: Virtual Bodies in Cybernetics, Literature, and Informatics; The University of Chicago Press: Chicago, IL, USA; London, UK, 1999. [Google Scholar]
- Bateson, G. Steps to an Ecology of Mind; Jason Aronson Inc.: Northvale, NJ, USA; London, UK, 1972. [Google Scholar]
- Pask, G. An Approach to Cybernetics; Hutchinson & Co., Ltd.: London, UK, 1961. [Google Scholar]
- Del Moro, L.; Ruzzante, B.; Magarini, M.; Gentili, P.L.; Rampioni, G.; Roli, A.; Damiano, L.; Stano, P. Chemical Neural Networks and Semantic Information Investigated Through Synthetic Cells. In Proceedings of the Artificial Life and Evolutionary Computation (WIVACE) 2022, Gaeta, Italy, 14–16 September 2022; De Stefano, C., Fontanella, F., Vanneschi, L., Eds.; Communications in Computer and Information Science. Springer Nature Switzerland: Cham, Switzerland, 2023; Volume 1780, pp. 27–39. [Google Scholar] [CrossRef]
- Lan, Q.; Wen, D.; Zhang, Z.; Zeng, Q.; Chen, X.; Popovski, P.; Huang, K. What is semantic communication? A view on conveying meaning in the era of machine intelligence. J. Commun. Inf. Netw. 2021, 6, 336–371. [Google Scholar] [CrossRef]
- Turing, A.M.I. Computing Machinery and Intelligence. Mind 1950, LIX, 433–460. [Google Scholar] [CrossRef]
- Reza, F.M. An Introduction to Information Theory; Dover Publications, Inc.: New York, NY, USA, 1994. [Google Scholar]
- Wiener, N. Cybernetics or, Control and Communication in the Animal and the Machine, 1st ed.; MIT Press: Cambridge, MA, USA, 1948. [Google Scholar]
- Ashby, W.R. An Introduction to Cybernetics; John Wiley & Sons Inc.: New York, NY, USA, 1956. [Google Scholar]
- von Foerster, H. Understanding Understanding: Essays on Cybernetics and Cognition; Springer: New York, NY, USA, 2003. [Google Scholar]
- Chen, G.; Levin, R.; Landau, S.; Kaduri, M.; Adir, O.; Ianovici, I.; Krinsky, N.; Doppelt-Flikshtain, O.; Shklover, J.; Shainsky-Roitman, J.; et al. Implanted synthetic cells trigger tissue angiogenesis through de novo production of recombinant growth factors. Proc. Natl. Acad. Sci. USA 2022, 119, e2207525119. [Google Scholar] [CrossRef]
- Bolinger, P.Y.; Stamou, D.; Vogel, H. An integrated self-assembled nanofluidic system for controlled biological chemistries. Angew. Chem. Int. Ed. Engl. 2008, 47, 5544–5549. [Google Scholar] [CrossRef]
- Muñuzuri, A.P.; Pérez-Mercader, J. Unified representation of Life’s basic properties by a 3-species Stochastic Cubic Autocatalytic Reaction-Diffusion system of equations. Phys. Life Rev. 2022, 41, 64–83. [Google Scholar] [CrossRef]
- Epstein, I.R. Coupled chemical oscillators and emergent system properties. Chem. Commun. 2014, 50, 10758–10767. [Google Scholar] [CrossRef]
- Taylor, A.F.; Tinsley, M.R.; Showalter, K. Insights into collective cell behaviour from populations of coupled chemical oscillators. Phys. Chem. Chem. Phys. 2015, 17, 20047–20055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gentili, P.L.; Micheau, J.C. Light and chemical oscillations: Review and perspectives. J. Photochem. Photobiol. C Photochem. Rev. 2020, 43, 100321. [Google Scholar] [CrossRef]
- Liu, Y.; Pérez-Mercader, J.; Kiss, I.Z. Synchronization of Belousov–Zhabotinsky oscillators with electrochemical coupling in a spontaneous process. Chaos 2022, 32, 093128. [Google Scholar] [CrossRef] [PubMed]
- Proskurkin, I.S.; Smelov, P.S.; Vanag, V.K. Experimental verification of an opto-chemical “neurocomputer”. Phys. Chem. Chem. Phys. 2020, 22, 19359–19367. [Google Scholar] [CrossRef]
- Gentili, P.L.; Giubila, M.S.; Germani, R.; Romani, A.; Nicoziani, A.; Spalletti, A.; Heron, B.M. Optical Communication among Oscillatory Reactions and Photo-Excitable Systems: UV and Visible Radiation Can Synchronize Artificial Neuron Models. Angew. Chem. Int. Ed. 2017, 56, 7535–7540. [Google Scholar] [CrossRef]
- Vanwiggeren, G.D.; Roy, R. Chaotic communication using time-delayed optical systems. Int. J. Bifurc. Chaos 1999, 9, 2129–2156. [Google Scholar] [CrossRef] [Green Version]
- Gentili, P.L.; Dolnik, M.; Epstein, I.R. “Photochemical Oscillator”: Colored Hydrodynamic Oscillations and Waves in a Photochromic System. J. Phys. Chem. C 2014, 118, 598–608. [Google Scholar] [CrossRef]
- Gentili, P.L.; Giubila, M.S.; Heron, B.M. Processing Binary and Fuzzy Logic by Chaotic Time Series Generated by a Hydrodynamic Photochemical Oscillator. ChemPhysChem 2017, 18, 1831–1841. [Google Scholar] [CrossRef]
- Hayashi, K.; Gotoda, H.; Gentili, P.L. Probing and exploiting the chaotic dynamics of a hydrodynamic photochemical oscillator to implement all the basic binary logic functions. Chaos 2016, 26, 053102. [Google Scholar] [CrossRef]
- Gentili, P.L.; Giubila, M.S.; Germani, R.; Heron, B.M. Photochromic and luminescent compounds as artificial neuron models. Dye. Pigment. 2018, 156, 149–159. [Google Scholar] [CrossRef] [Green Version]
- Gentili, P.L. Photochromic and luminescent materials for the development of Chemical Artificial Intelligence. Dye. Pigment. 2022, 205, 110547. [Google Scholar] [CrossRef]
- Bartolomei, B.; Heron, B.M.; Gentili, P.L. A contribution to neuromorphic engineering: Neuromodulation implemented through photochromic compounds maintained out of equilibrium by UV-visible radiation. Rend. Lincei Sci. Fis. Nat. 2020, 31, 39–52. [Google Scholar] [CrossRef] [Green Version]
- Katz, P.S.; Calin-Jageman, R.J. Neuromodulation. In Encyclopedia of Neuroscience; Squire, L.R., Ed.; Academic Press: Oxford, UK, 2009; pp. 497–503. [Google Scholar] [CrossRef]
- Mallphanov, I.L.; Vanag, V.K. Distance dependent types of coupling of chemical micro-oscillators immersed in a water-in-oil microemulsion. Phys. Chem. Chem. Phys. 2021, 23, 9130–9138. [Google Scholar] [CrossRef]
- Tomasi, R.; Noël, J.M.; Zenati, A.; Ristori, S.; Rossi, F.; Cabuil, V.; Kanoufi, F.; Abou-Hassan, A. Chemical communication between liposomes encapsulating a chemical oscillatory reaction. Chem. Sci. 2014, 5, 1854–1859. [Google Scholar] [CrossRef]
- Draper, T.C.; Phillips, N.; Weerasekera, R.; Mayne, R.; Fullarton, C.; de Lacy Costello, B.P.J.; Adamatzky, A. Contactless sensing of liquid marbles for detection, characterisation & computing. Lab Chip 2020, 20, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Quaglia, G.; Bartolomei, B.; Gentili, P.L.; Latterini, L. UV-Visible radiation modulation abilities of photon up-converting nanocapsules integrated with an oscillatory reaction. J. Mater. Chem. C 2022, 10, 9073–9080. [Google Scholar] [CrossRef]
- Bray, D. Protein molecules as computational elements in living cells. Nature 1995, 376, 307–312. [Google Scholar] [CrossRef]
- Jakob, U.; Kriwacki, R.; Uversky, V.N. Conditionally and Transiently Disordered Proteins: Awakening Cryptic Disorder To Regulate Protein Function. Chem. Rev. 2014, 114, 6779–6805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gentili, P.L. The Fuzziness of the Molecular World and Its Perspectives. Molecules 2018, 23, 2074. [Google Scholar] [CrossRef] [Green Version]
- Zadeh, L.A. Outline of a New Approach to the Analysis of Complex Systems and Decision Processes. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 28–44. [Google Scholar] [CrossRef] [Green Version]
- Gentili, P.L. Establishing a New Link between Fuzzy Logic, Neuroscience, and Quantum Mechanics through Bayesian Probability: Perspectives in Artificial Intelligence and Unconventional Computing. Molecules 2021, 26, 5987. [Google Scholar] [CrossRef] [PubMed]
- Damiano, L.; Dumouchel, P.G. Emotions in Relation. Epistemological and Ethical Scaffolding for Mixed Human-Robot Social Ecologies. J. Philos. Stud. 2020, 13, 181–206. [Google Scholar]
- Fong, T.; Nourbakhsh, I.; Dautenhahn, K. A survey of socially interactive robots. Robot. Auton. Syst. 2003, 42, 143–166. [Google Scholar] [CrossRef] [Green Version]
- Dautenhahn, K. Socially intelligent robots: Dimensions of human-robot interaction. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2007, 362, 679–704. [Google Scholar] [CrossRef] [Green Version]
- Daily, S.B.; James, M.T.; Cherry, D.; Porter, J.J., III; Darnell, S.S.; Isaac, J.; Roy, T. Affective Computing: Historical Foundations, Current Applications, and Future Trends. In Emotions and Affect in Human Factors and Human-Computer Interaction; Jeon, M., Ed.; Academic Press: London, UK, 2017; pp. 213–231. [Google Scholar]
- Miłkowski, M.; Clowes, R.; Rucińska, Z.; Przegalińska, A.; Zawidzki, T.; Krueger, J.; Gies, A.; McGann, M.; Afeltowicz, Ł.; Wachowski, W.; et al. From Wide Cognition to Mechanisms: A Silent Revolution. Front. Psychol. 2018, 9, 2393. [Google Scholar] [CrossRef] [Green Version]
- Damiano, L. Homes as human–robot ecologies: An epistemological inquiry on the “domestication” of robots. In The Home in the Digital Age; Routledge: Abingdon, UK, 2021. [Google Scholar]
- Dumouchel, P.; Damiano, L. Living with Robots; Harvard University Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Kolisis, N.; Kolisis, F. Synthetic Biology: Old and New Dilemmas-The Case of Artificial Life. BioTech 2021, 10, 16. [Google Scholar] [CrossRef]
- Mackelprang, R.; Aurand, E.R.; Bovenberg, R.A.L.; Brink, K.R.; Charo, R.A.; Delborne, J.A.; Diggans, J.; Ellington, A.D.; Fortman, J.L.C.; Isaacs, F.J.; et al. Guiding Ethical Principles in Engineering Biology Research. ACS Synth. Biol. 2021, 10, 907–910. [Google Scholar] [CrossRef]
- Gómez-Tatay, L.; Hernández-Andreu, J.M. Biosafety and biosecurity in Synthetic Biology: A review. Crit. Rev. Environ. Sci. Technol. 2019, 49, 1587–1621. [Google Scholar] [CrossRef]
- Filipczak, N.; Pan, J.; Yalamarty, S.S.K.; Torchilin, V.P. Recent advancements in liposome technology. Adv. Drug Deliv. Rev. 2020, 156, 4–22. [Google Scholar] [CrossRef]
- Wang, S.; Chen, Y.; Guo, J.; Huang, Q. Liposomes for Tumor Targeted Therapy: A Review. Int. J. Mol. Sci. 2023, 24, 2643. [Google Scholar] [CrossRef]





| Feature | Telecommunication | Molecular Communication |
|---|---|---|
| Signal | Electromagnetic | Chemical |
| Speed | Fast | Slow |
| Distance | Long | Short |
| Media | Air, cables | Aqueous (mainly) |
| Transmission | Accurate | Stochastic |
| Research Area | Scopes |
|---|---|
| Origin of Life, Systems Chemistry | Building models of primitive cells (protocells) in order to shed light on the non-life-to-life transition; investigating cell-like structures made of primitive chemicals and functioning by primitive mechanisms; determining the minimal complexity of life; identifying the essential components and processes for the onset of life on Earth; understanding the emergence of life; investigating fundamental mechanisms, such as out-of-equilibrium dynamics, self-replication and self-reproduction, oscillations, confinement, nonlinearity, etc. |
| Sciences of the Artificial, Artificial Life, Theoretical Biology | Studying autonomy, agency, autopoiesis, self-organization, complexity, evolution, emergence of meaning, biosemiotics, etc., by means of physical or virtual cell models; understanding and comparing wetware models of life and cognition to hardware and software models (robotics and AI); exploring chemical AI, embodied AI, chemical robotics, autonomy, agency, communication and information theories, unconventional computing, cognition, enaction, embodiment, emergence, etc. |
| Molecular Communications, IoBNT | Building SCs as bio-nanoelements (devices) operating in bio-nanonetworks via molecular communication; engineering manipulation and transfer of chemical information; investigating communication and information theories, cybernetic aspects of molecular systems and syntactic and semantic information; constructing systems with sensing, control, actuation abilities, etc. |
| Biochemistry, Biophysics, Synthetic Biology, Biotechnological Applications | Building and using cell-like systems as an experimental platform to investigate complex biochemical or biophysical phenomena (i.e., reconstructing the target phenomenon in a simplified, and thus easier to study, context); exploring possible uses in nanomedicine, smart drug delivery, biosensing, bioproduction, nanoreactors, enzyme immobilization, tissue engineering, bioassays, etc. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stano, P.; Gentili, P.L.; Damiano, L.; Magarini, M. A Role for Bottom-Up Synthetic Cells in the Internet of Bio-Nano Things? Molecules 2023, 28, 5564. https://doi.org/10.3390/molecules28145564
Stano P, Gentili PL, Damiano L, Magarini M. A Role for Bottom-Up Synthetic Cells in the Internet of Bio-Nano Things? Molecules. 2023; 28(14):5564. https://doi.org/10.3390/molecules28145564
Chicago/Turabian StyleStano, Pasquale, Pier Luigi Gentili, Luisa Damiano, and Maurizio Magarini. 2023. "A Role for Bottom-Up Synthetic Cells in the Internet of Bio-Nano Things?" Molecules 28, no. 14: 5564. https://doi.org/10.3390/molecules28145564
APA StyleStano, P., Gentili, P. L., Damiano, L., & Magarini, M. (2023). A Role for Bottom-Up Synthetic Cells in the Internet of Bio-Nano Things? Molecules, 28(14), 5564. https://doi.org/10.3390/molecules28145564