Natural and Synthetic Anticancer Epidrugs Targeting the Epigenetic Integrator UHRF1
Abstract
:1. Introduction
2. Structure and Function of UHRF1
2.1. Structure of UHRF1
2.1.1. Ubiquitin-like Domain (UBL)
2.1.2. Tandem Tudor Domain (TTD)
2.1.3. Plant Homeodomain (PHD)
2.1.4. Set and Ring-Associated (SRA) Domain
2.1.5. RING Domain
2.2. Functions of UHRF1
3. UHRF1 Expression and Its Regulation
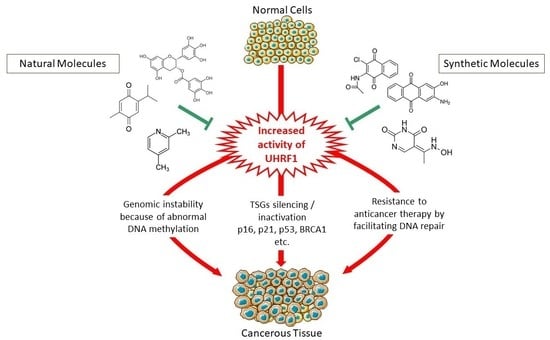
4. Why Targeting UHRF1 Is an Interesting Direction to Promote
5. Synthetic Molecules Targeting UHRF1
5.1. Molecules Directly Targeting UHRF1 Protein
5.1.1. Targeting the SRA Domain of UHRF1
5.1.2. Targeting the TTD Domain of UHRF1
5.2. Synthetic Drugs Affecting UHRF1 at the Transcriptional Level and/or Protein Level
5.3. Limitations of Synthetic Compounds Targeting UHRF1
| Molecules Directly Targeting UHRF1 Protein | ||||
|---|---|---|---|---|
| Drug | Cancers | Mechanism | Function | Reference |
| Targeting the SRA domain of UHRF1 | ||||
| NSC232003 | Glioma | Binds to 5-mC pocket of UHRF1 and inhibits its function | Impaired interaction of UHRF1/DNMT1 Global hypomethylation | [100] |
| UM63 | Cervical | Inhibits recognition and base flipping of 5-mC. Impairs UHRF1/DNMT1 interaction | Reduced global methylation levels | [98] |
| AMSA2 MPB7 | Cervical Melanoma Breast ductal carcinoma | Inhibits UHRF1-SRA-mediated base flipping and recruitment of DNMT1 at replication foci | Reduced global methylation levels and induced apoptosis of cancer cells | [102] |
| LOPAC compounds (mitoxantrone, doxorubicin, idarubicin, pixantrone, and daunorubicin) | Prostate | Inhibits SRA/HM DNA binding | Global demethylation, synergistic cytotoxic effect on tumor cells in combination with decitabine | [103] |
| Targeting the TTD domain of UHRF1 | ||||
| BPC | Binds with TTD and favors UHRF1 open conformation | Impaired interaction of UHRF1 with H3K9me3 | [104] | |
| NV01 and NV03 | Binds with TTD domain of UHRF1 | Disrupts UHRF1–H3K9me3 interaction | [105] | |
| 5A-DMP | Colorectal | Inhibition of TTD interaction with LIG1 | Inhibits interaction of TTD domain with LIG1, which is crucial for maintenance of DNA methylation | [106] |
| Synthetic drugs affecting UHRF1 at the transcriptional level and/or protein level | ||||
| LY294002, GF109203X, PD98059, AG490, and genistein. PD0325901 in combination with CHIR99021 | Breast, liver, and ALL | Releases the transcription factor E2F to regulate UHRF1 expression | Inhibition of cellular proliferation and colony formation, cell cycle arrest, and transcriptional regulation of UHRF1 and DNMT1 | [109,110] |
| Dihydroartemisinin | Prostate | Down-regulation of UHRF1 and DNMT1, up-regulation of TSG p16 | Cell cycle arrest at G1/S, apoptosis, and inhibition of cell proliferation and metastasis | [111,112,113,114] |
| Torin-2 | HCC, colorectal, SCLC, NSCLC, and ESCC | Inhibition of PI3K/Akt/mTOR signaling pathway that regulates UHRF1 expression | Inhibition of migration, proliferation, and EMT, and apoptosis induction | [118,119,120,121,122,123] |
6. Natural Compounds Targeting UHRF1
6.1. Natural Compounds Directly Targeting UHRF1 Protein
6.2. Natural Compounds Affecting UHRF1 Gene Expression
6.2.1. Plant Extracts
6.2.2. Purified Plant Drugs
6.3. Natural Compounds Targeting UHRF1 at Gene and Protein Levels
Natural Compounds Purified from Bacteria
6.4. Limitations of Natural Compounds Targeting UHRF1
| Natural Compounds Directly Targeting UHRF1 Protein | ||||
|---|---|---|---|---|
| Drug | Cancers | Mechanism | Function | Reference |
| Chicoric acid | Colorectal | Binds with SRA domain and inhibits its activity | Reduces methylation levels | [126] |
| Berberine | Multiple myeloma | Binds with TTD and PHD domain of UHRF1 and induces ubiquitination-mediated degradation of UHRF1, activates p16INK4A and p73 | Inhibits cell growth and cytotoxic in multiple myeloma cells | [127] |
| 2,4-Lutidine | Inhibits binding of TTD with H3K9me3 | Inhibits H3K9me3 mark recognition by TTD and may induce the expression of TSGs | [128] | |
| Natural Compounds Affecting UHRF1 Gene Expression | ||||
| Plant extracts | ||||
| Rhaponticum carthamoides root extract | Glioma | Down-regulation of UHRF1 and DNMT1 mRNA levels, cleavage of PARP, and inhibition of PARP synthesis | Apoptosis | [130] |
| Leonurus sibiricus root extract | Glioma | In combination with AtPAP1, transcription factor induces down-regulation of UHRF1 and DNMT1, cleavage of PARP, and an increase in γH2A.X. | DNA damage | [131] |
| Vaccinium myrtillus, Bilberry extract, Aronia melanocarpa | B cell chronic lymphocytic leukemia and Jurkat cells | Down-regulation of UHRF1 and Cyclin B1, activation of p73 and caspase-3 expression, inhibition of Bcl-2, and dephosphorylation of Akt and Bad | Anti-tumor, anti-angiogenesis, anti-proliferative, and cell cycle arrest in G2/M phase | [132,134,135,136] |
| Maritime pine tannin extract | Cervical and osteosarcoma | Down-regulation of UHRF1 and DNMT1, induces expression of p73 and caspase-3, cleavage of PARP, and down-regulation of pro-apoptotic Bcl-2 | Anti-proliferative, G2/M phase growth arrest, global hypomethylation, and apoptosis | [137] |
| Red wine-derived polyphenols | Leukemia and C26 carcinoma | Down-regulation of UHRF1 and induces expression of TSGs: p16, p53, p73, PAX1, and caspase-3 protein | Reduced cell viability, cell cycle arrest, apoptosis, inhibition of proliferation, and angiogenesis | [133,138,139] |
| Limoniastrum guyonianum | Cervical | Down-regulates UHRF1 and DNMT1, activates the expression of TSG p16INK4A | Cell cycle arrest at G2/M phase, apoptosis, and global hypomethylation | [140] |
| Purified plant drugs | ||||
| TIT3 | HCC | Down-regulation of UHRF1, DNMT1, HDAC7, and DNA repair genes, and up-regulation of proapoptotic genes | Anti-proliferative and proapoptotic effect | [142] |
| Luteolin | Colorectal | Down-regulation of UHRF1 and DNMT1, re-expression of TSG p16INK4A, and PARP cleavage. | Antiproliferative, cell cycle arrest, and apoptosis | [140,143] |
| EGCG | ALL | ROS-dependent down-regulation of UHRF1 and DNMT1, and re-expression of TSGs: p73 and p16INK4A | Cell cycle arrest and apoptosis | [144] |
| Shikonin (alone or in combination with melatonin) | NSCLC, breast, pancreatic, cervical, and osteosarcoma | Down-regulation of UHRF1, re-expression of TSG p16INK4A, p73 and caspase-3-dependent apoptosis, and oxidative stress mediated-apoptosis | Cell cycle arrest, apoptosis, and autophagy | [145,146,147] |
| Hinokitiol | Colon | Down-regulation of UHRF1 and DNMT1, and induces expression of TET1 protein and TSGs involved in cell proliferation | Anti-proliferative, apoptosis, and demethylation | [148] |
| Emodin (alone or in combination with doxorubicin) | Lymphoma Raji cells | Inhibition of UHRF1 expression and activation of caspase-3, caspase-9, and PARP | Growth arrest, apoptosis, reduced cell viability, and enhanced tumor cell sensitivity to doxorubicin | [149] |
| Diosgenin | Prostate | Ubiquitin-mediated degradation of UHRF1, down-regulation of DNA methylation, and activation of TSG: p16, p21, and LXN | Cell cycle arrest, senescence, and inhibition of cellular proliferation and xenograft tumor growth | [150] |
| Natural Compounds Targeting UHRF1 at Gene and Protein Level | ||||
| Thymoquinone | Renal, colorectal, osteosarcoma, astrocytoma, ovarian adenocarcinoma, cervical, and breast | Down-regulation of UHRF1/DNMT1/HDAC1/G9a/USP7 and re-expression of TSGs: p16 and p73. Induces expression of caspase-3 | Anti-proliferative, cell cycle arrest, and p53-dependent and p53-independent apoptosis | [151,152,153,154,155,156,157,158,159] |
| Naphthazarin | Breast and t(1;19)-pre-B-cell acute lymphoblastic leukemia | Down-regulation of UHRF1/DNMT1/HDAC1, induces p53-dependent p21 expression, and inhibition of ROR1 expression | Cell cycle arrest, apoptosis, enhances radio sensitivity of MCF-7 breast cancer cells, and reduces ALL cell viability | [160,162] |
| Plumbagin (alone or in combination with cisplatin) | Cervical | Down-regulation of UHRF1 at transcript and protein levels and down-regulation of Akt-1, caspase 9, and PARP1 | Inhibition of metastasis and proliferation, and synergistic apoptosis in combination with cisplatin | [163] |
| Curcumin | Melanoma and glioblastoma | Targets PDE1 enzyme, down-regulation of UHRF1, DNMT1, and cyclin A, activation of p21, p27, and PAX1, and ROS generation | Anti-proliferative, cell cycle arrest, reduced viability of glioblastoma cells in combination with sodium butyrate, and apoptosis | [139,164,165] |
| Natural compounds purified from bacteria | ||||
| Anisomycin | ALL | Down-regulation of UHRF1 and activation of TSGs: p21, p27, and p53 | Cell cycle arrest at G2/M phase | [166] |
| Mithramycin | Malignant pleural mesotheliomas | Down-regulation of UHRF1 | Targeting DNA methylation | [167] |
7. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, W.; Henneberg, M. Cancer incidence increasing globally: The role of relaxed natural selection. Evol. Appl. 2017, 11, 140–152. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 2008, 358, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Dupont, C.; Armant, D.R.; Brenner, C.A. Epigenetics: Definition, Mechanisms and Clinical Perspective; Thieme Medical Publishers: New York, NY, USA, 2009; Volume 27, pp. 351–357. [Google Scholar]
- Lund, A.H.; van Lohuizen, M. Epigenetics and Cancer. Genes Dev. 2004, 18, 2315–2335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammad, H.P.; Barbash, O.; Creasy, C.L. Targeting epigenetic modifications in cancer therapy: Erasing the roadmap to cancer. Nat. Med. 2019, 25, 403–418. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, K.; Das, V.; Vyas, P.; Hajdúch, M. Nucleosidic DNA demethylating epigenetic drugs—A comprehensive review from discovery to clinic. Pharmacol. Ther. 2018, 188, 45–79. [Google Scholar] [CrossRef]
- Cappellacci, L.; Perinelli, D.R.; Maggi, F.; Grifantini, M.; Petrelli, R. Recent Progress in Histone Deacetylase Inhibitors as Anticancer Agents. Curr. Med. Chem. 2020, 27, 2449–2493. [Google Scholar] [CrossRef]
- Hopfner, R.; Mousli, M.; Garnier, J.-M.; Redon, R.; du Manoir, S.; Chatton, B.; Ghyselinck, N.; Oudet, P.; Bronner, C. Genomic structure and chromosomal mapping of the gene coding for ICBP90, a protein involved in the regulation of the topoisomerase IIα gene expression. Gene 2001, 266, 15–23. [Google Scholar] [CrossRef]
- Hopfner, R.; Mousli, M.; Jeltsch, J.M.; Voulgaris, A.; Lutz, Y.; Marin, C.; Bellocq, J.P.; Oudet, P.; Bronner, C. ICBP90, a Novel Human CCAAT Binding Protein, Involved in the Regulation of Topoisomerase IIalpha Expression. Cancer Res. 2000, 60, 121–128. [Google Scholar]
- Bronner, C.; Achour, M.; Arima, Y.; Chataigneau, T.; Saya, H.; Schini-Kerth, V.B. The UHRF family: Oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol. Ther. 2007, 115, 419–434. [Google Scholar] [CrossRef] [PubMed]
- Mancini, M.; Magnani, E.; Macchi, F.; Bonapace, I.M. The multi-functionality of UHRF1: Epigenome maintenance and preservation of genome integrity. Nucleic Acids Res. 2021, 49, 6053–6068. [Google Scholar] [CrossRef] [PubMed]
- Bronner, C.; Krifa, M.; Mousli, M. Increasing role of UHRF1 in the reading and inheritance of the epigenetic code as well as in tumorogenesis. Biochem. Pharmacol. 2013, 86, 1643–1649. [Google Scholar] [CrossRef] [PubMed]
- Bronner, C.; Alhosin, M.; Hamiche, A.; Mousli, M. Coordinated Dialogue between UHRF1 and DNMT1 to Ensure Faithful Inheritance of Methylated DNA Patterns. Genes 2019, 10, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smets, M.; Link, S.; Wolf, P.; Schneider, K.; Solis, V.; Ryan, J.; Meilinger, D.; Qin, W.; Leonhardt, H. DNMT1 mutations found in HSANIE patients affect interaction with UHRF1 and neuronal differentiation. Hum. Mol. Genet. 2017, 26, 1522–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unoki, M.; Sasaki, H. The UHRF protein family in epigenetics, development, and carcinogenesis. Proc. Jpn. Acad. Ser. B 2022, 98, 401–415. [Google Scholar] [CrossRef] [PubMed]
- DaRosa, P.A.; Harrison, J.S.; Zelter, A.; Davis, T.N.; Brzovic, P.; Kuhlman, B.; Klevit, R.E. A Bifunctional Role for the UHRF1 UBL Domain in the Control of Hemi-methylated DNA-Dependent Histone Ubiquitylation. Mol. Cell 2018, 72, 753–765.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foster, B.M.; Stolz, P.; Mulholland, C.B.; Montoya, A.; Kramer, H.; Bultmann, S.; Bartke, T. Critical Role of the UBL Domain in Stimulating the E3 Ubiquitin Ligase Activity of UHRF1 toward Chromatin. Mol. Cell 2018, 72, 739–752.e9. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Wang, L.; Du, Y.; Xie, S.; Yang, X.; Lian, F.; Zhou, Z.; Qian, C. Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation. Nucleic Acids Res. 2018, 46, 3218–3231. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.; Wang, G.G. Tudor: A versatile family of histone methylation ‘readers’. Trends Biochem. Sci. 2013, 38, 546–555. [Google Scholar] [CrossRef] [Green Version]
- Nady, N.; Lemak, A.; Walker, J.R.; Avvakumov, G.V.; Kareta, M.S.; Achour, M.; Xue, S.; Duan, S.; Allali-Hassani, A.; Zuo, X.; et al. Recognition of Multivalent Histone States Associated with Heterochromatin by UHRF1 Protein. J. Biol. Chem. 2011, 286, 24300–24311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferry, L.; Fournier, A.; Tsusaka, T.; Adelmant, G.; Shimazu, T.; Matano, S.; Kirsh, O.; Amouroux, R.; Dohmae, N.; Suzuki, T.; et al. Methylation of DNA Ligase 1 by G9a/GLP Recruits UHRF1 to Replicating DNA and Regulates DNA Methylation. Mol. Cell 2017, 67, 550–565.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaughan, R.M.; Kupai, A.; Foley, C.A.; Sagum, C.A.; Tibben, B.M.; Eden, H.E.; Tiedemann, R.L.; Berryhill, C.A.; Patel, V.; Shaw, K.M.; et al. The histone and non-histone methyllysine reader activities of the UHRF1 tandem Tudor domain are dispensable for the propagation of aberrant DNA methylation patterning in cancer cells. Epigenetics Chromatin 2020, 13, 44. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, A. Novel Insights into Peptide Binding and Conformational Dynamics of UHRF1. Structure 2019, 27, 408–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kori, S.; Ferry, L.; Matano, S.; Jimenji, T.; Kodera, N.; Tsusaka, T.; Matsumura, R.; Oda, T.; Sato, M.; Dohmae, N.; et al. Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation. Structure 2019, 27, 485–496.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, J.; Cheng, J.; Wang, J.; Zhang, Q.; Liu, M.; Gong, R.; Wang, P.; Zhang, X.; Feng, Y.; Lan, W.; et al. Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition. Nat. Commun. 2016, 7, 11197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, L.; Tan, X.-F.; Zhang, S.; Wu, T.; Zhang, Z.-M.; Ai, H.-W.; Song, J. An Intramolecular Interaction of UHRF1 Reveals Dual Control for Its Histone Association. Structure 2018, 26, 304–311.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.-M.; Rothbart, S.B.; Allison, D.F.; Cai, Q.; Harrison, J.S.; Li, L.; Wang, Y.; Strahl, B.D.; Wang, G.G.; Song, J. An Allosteric Interaction Links USP7 to Deubiquitination and Chromatin Targeting of UHRF1. Cell Rep. 2015, 12, 1400–1406. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; Li, Z.; Wang, P.; Lin, Y.; Xu, Y. Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2. Cell Res. 2011, 21, 1374–1378. [Google Scholar] [CrossRef]
- Rajakumara, E.; Wang, Z.; Ma, H.; Hu, L.; Chen, H.; Lin, Y.; Guo, R.; Wu, F.; Li, H.; Lan, F.; et al. PHD Finger Recognition of Unmodified Histone H3R2 Links UHRF1 to Regulation of Euchromatic Gene Expression. Mol. Cell 2011, 43, 275–284. [Google Scholar] [CrossRef] [Green Version]
- Hata, K.; Kobayashi, N.; Sugimura, K.; Qin, W.; Haxholli, D.; Chiba, Y.; Yoshimi, S.; Hayashi, G.; Onoda, H.; Ikegami, T.; et al. Structural basis for the unique multifaceted interaction of DPPA3 with the UHRF1 PHD finger. Nucleic Acids Res. 2022, 50, 12527–12542. [Google Scholar] [CrossRef]
- Avvakumov, G.V.; Walker, J.R.; Xue, S.; Li, Y.; Duan, S.; Bronner, C.; Arrowsmith, C.H.; Dhe-Paganon, S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 2008, 455, 822–825. [Google Scholar] [CrossRef] [PubMed]
- Arita, K.; Ariyoshi, M.; Tochio, H.; Nakamura, Y.; Shirakawa, M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 2008, 455, 818–821. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Horton, J.R.; Zhang, X.; Bostick, M.; Jacobsen, S.E.; Cheng, X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 2008, 455, 826–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Wu, K.; Chen, S.; Jiang, S.; Chen, Y.; Duan, C. UHRF2 Promotes Hepatocellular Carcinoma Progression by Upregu-lating ErbB3/Ras/Raf Signaling Pathway. Int. J. Med. Sci. 2021, 18, 3097. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Duan, Q.; Zeng, Z.; Zhao, J.; Lu, J.; Sun, J.; Zhang, J.; Siwko, S.; Wong, J.; Shi, T.; et al. UHRF2 promotes intestinal tumorigenesis through stabilization of TCF4 mediated Wnt/β-catenin signaling. Int. J. Cancer 2020, 147, 2239–2252. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Xiong, J.; Wang, M.; Yang, N.; Wong, J.; Zhu, B.; Xu, R.-M. Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2. Mol. Cell 2014, 54, 879–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenkins, Y.; Markovtsov, V.; Lang, W.; Sharma, P.; Pearsall, D.; Warner, J.; Franci, C.; Huang, B.; Huang, J.; Yam, G.C.; et al. Critical Role of the Ubiquitin Ligase Activity of UHRF1, a Nuclear RING Finger Protein, in Tumor Cell Growth. Mol. Biol. Cell 2005, 16, 5621–5629. [Google Scholar] [CrossRef]
- Du, Z.; Song, J.; Wang, Y.; Zhao, Y.; Guda, K.; Yang, S.; Kao, H.-Y.; Xu, Y.; Willis, J.; Markowitz, S.D.; et al. DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination. Sci. Signal. 2010, 3, ra80. [Google Scholar] [CrossRef] [Green Version]
- Bostick, M.; Kim, J.K.; Estève, P.-O.; Clark, A.; Pradhan, S.; Jacobsen, S.E. UHRF1 Plays a Role in Maintaining DNA Methylation in Mammalian Cells. Science 2007, 317, 1760–1764. [Google Scholar] [CrossRef] [Green Version]
- Sharif, J.; Muto, M.; Takebayashi, S.-I.; Suetake, I.; Iwamatsu, A.; Endo, T.A.; Shinga, J.; Mizutani-Koseki, Y.; Toyoda, T.; Okamura, K.; et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 2007, 450, 908–912. [Google Scholar] [CrossRef] [PubMed]
- Karagianni, P.; Amazit, L.; Qin, J.; Wong, J. ICBP90, a Novel Methyl K9 H3 Binding Protein Linking Protein Ubiquitination with Heterochromatin Formation. Mol. Cell. Biol. 2008, 28, 705–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothbart, S.B.; Krajewski, K.; Nady, N.; Tempel, W.; Xue, S.; Badeaux, A.I.; Barsyte-Lovejoy, D.; Martinez, J.Y.; Bedford, M.T.; Fuchs, S.M.; et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat. Struct. Mol. Biol. 2012, 19, 1155–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rottach, A.; Frauer, C.; Pichler, G.; Bonapace, I.M.; Spada, F.; Leonhardt, H. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res. 2009, 38, 1796–1804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gelato, K.A.; Tauber, M.; Ong, M.S.; Winter, S.; Hiragami-Hamada, K.; Sindlinger, J.; Lemak, A.; Bultsma, Y.; Houliston, S.; Schwarzer, D.; et al. Accessibility of Different Histone H3-Binding Domains of UHRF1 Is Allosterically Regulated by Phosphatidylinositol 5-Phosphate. Mol. Cell 2014, 54, 905–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mistry, H.; Tamblyn, L.; Butt, H.; Sisgoreo, D.; Gracias, A.; Larin, M.; Gopalakrishnan, K.; Hande, M.P.; McPherson, J.P. UHRF1 is a genome caretaker that facilitates the DNA damage response to γ-irradiation. Genome Integr. 2010, 1, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Wang, Y.; Zhang, F.; Sun, G.; Li, C.; Jing, S.; Liu, Q.; Cheng, Y. Inhibiting UHRF1 expression enhances radiosensitivity in human esophageal squamous cell carcinoma. Mol. Biol. Rep. 2013, 40, 5225–5235. [Google Scholar] [CrossRef]
- Tian, Y.; Paramasivam, M.; Ghosal, G.; Chen, D.; Shen, X.; Huang, Y.; Akhter, S.; Legerski, R.; Chen, J.; Seidman, M.M.; et al. UHRF1 Contributes to DNA Damage Repair as a Lesion Recognition Factor and Nuclease Scaffold. Cell Rep. 2015, 10, 1957–1966. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.-C.; Zhan, B.; Yoshikawa, Y.; Haas, W.; Gygi, S.P.; Cohn, M.A. UHRF1 Is a Sensor for DNA Interstrand Crosslinks and Recruits FANCD2 to Initiate the Fanconi Anemia Pathway. Cell Rep. 2015, 10, 1947–1956. [Google Scholar] [CrossRef] [Green Version]
- Motnenko, A.; Liang, C.-C.; Yang, D.; Lopez-Martinez, D.; Yoshikawa, Y.; Zhan, B.; Ward, K.E.; Tian, J.; Haas, W.; Spingardi, P.; et al. Identification of UHRF2 as a novel DNA interstrand crosslink sensor protein. PLoS Genet. 2018, 14, e1007643. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Liu, H.; Chen, Y.; Yang, X.; Wang, P.; Liu, T.; Deng, M.; Qin, B.; Correia, C.; Lee, S.; et al. A cell cycle-dependent BRCA1–UHRF1 cascade regulates DNA double-strand break repair pathway choice. Nat. Commun. 2016, 7, 10201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hahm, J.Y.; Kim, J.-Y.; Park, J.W.; Kang, J.-Y.; Kim, K.-B.; Kim, S.-R.; Cho, H.; Seo, S.-B. Methylation of UHRF1 by SET7 is essential for DNA double-strand break repair. Nucleic Acids Res. 2019, 47, 184–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hahm, J.Y.; Kang, J.-Y.; Park, J.W.; Jung, H.; Seo, S.-B. Methylated-UHRF1 and PARP1 Interaction Is Critical for Homologous Recombination. BMB Rep. 2020, 53, 112–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mousli, M.; Hopfner, R.; Abbady, A.-Q.; Monté, D.; Jeanblanc, M.; Oudet, P.; Louis, B.; Bronner, C. ICBP90 belongs to a new family of proteins with an expression that is deregulated in cancer cells. Br. J. Cancer 2003, 89, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Arima, Y.; Hirota, T.; Bronner, C.; Mousli, M.; Fujiwara, T.; Niwa, S.-I.; Ishikawa, H.; Saya, H. Down-regulation of nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage checkpoint signals contributes to cell cycle arrest at G1/S transition. Genes Cells 2004, 9, 131–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tien, A.L.; Senbanerjee, S.; Kulkarni, A.; Mudbhary, R.; Goudreau, B.; Ganesan, S.; Sadler, K.C.; Ukomadu, C. UHRF1 depletion causes a G2/M arrest, activation of DNA damage response and apoptosis. Biochem. J. 2011, 435, 175–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unoki, M.; Nishidate, T.; Nakamura, Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene 2004, 23, 7601–7610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.-A.; Platt, J.; Lee, J.W.; López-Giráldez, F.; Herbst, R.S.; Koo, J.S. E2F8 as a Novel Therapeutic Target for Lung Cancer. JNCI J. Natl. Cancer Inst. 2015, 107, djv151. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.-M.; Cheng, W.-L.; Liao, C.-J.; Chi, H.-C.; Lin, Y.-H.; Tseng, Y.-H.; Tsai, C.-Y.; Chen, C.-Y.; Lin, S.-L.; Chen, W.-J.; et al. Negative modulation of the epigenetic regulator, UHRF1, by thyroid hormone receptors suppresses liver cancer cell growth. Int. J. Cancer 2015, 137, 37–49. [Google Scholar] [CrossRef]
- Boukhari, A.; Alhosin, M.; Bronner, C.; Sagini, K.; Truchot, C.; Sick, E.; Schini-Kerth, V.B.; André, P.; Mély, Y.; Mousli, M.; et al. CD47 Activation-Induced UHRF1 Over-Expression Is Associated with Silencing of Tumor Suppressor Gene p16INK4A in Glioblastoma Cells. Anticancer Res. 2015, 35, 149. [Google Scholar]
- Cui, X.; Cui, Y.; Du, T.; Jiang, X.; Song, C.; Zhang, S.; Ma, C.; Liu, Y.; Ni, Q.; Gao, Y.; et al. SHMT2 Drives the Progression of Colorectal Cancer by Regulating UHRF1 Expression. Can. J. Gastroenterol. Hepatol. 2022, 2022, 3758697. [Google Scholar] [CrossRef]
- El Fersioui, Y.; Pinton, G.; Allaman-Pillet, N.; Schorderet, D.F. Hmx1 regulates urfh1 expression in the craniofacial region in zebrafish. PLoS ONE 2021, 16, e0245239. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhao, X.; Han, Y.; Lu, Y.; Shang, Y.; Liu, C.; Li, T.; Jin, Z.; Fan, D.; Wu, K. Regulation of UHRF1 by miR-146a/b modulates gastric cancer invasion and metastasis. FASEB J. 2013, 27, 4929–4939. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Yan, M.; Yu, T.; Ge, H.; Lin, H.; Li, J.; Liu, Y.; Geng, Q.; Zhu, M.; Liu, L.; et al. Quantitative Proteomic Analysis of the Metastasis-Inhibitory Mechanism of miR-193a-3p in Non-Small Cell Lung Cancer. Cell. Physiol. Biochem. 2015, 35, 1677–1688. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ma, H.; Inuzuka, H.; Diao, J.; Lan, F.; Shi, Y.G.; Wei, W.; Shi, Y. DNA Damage Regulates UHRF1 Stability via the SCFβ-TrCP E3 Ligase. Mol. Cell. Biol. 2013, 33, 1139–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, H.; Chen, H.; Guo, X.; Wang, Z.; Sowa, M.E.; Zheng, L.; Hu, S.; Zeng, P.; Guo, R.; Diao, J.; et al. M phase phosphorylation of the epigenetic regulator UHRF1 regulates its physical association with the deubiquitylase USP7 and stability. Proc. Natl. Acad. Sci. USA 2012, 109, 4828–4833. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, T.; Ashraf, W.; Ibrahim, A.; Zaayter, L.; Muller, C.D.; Hamiche, A.; Mély, Y.; Bronner, C.; Mousli, M. TIP60 governs the auto-ubiquitination of UHRF1 through USP7 dissociation from the UHRF1/USP7 complex. Int. J. Oncol. 2021, 59, 89. [Google Scholar] [CrossRef]
- Hong, Y.J.; Park, J.; Hahm, J.Y.; Kim, S.H.; Lee, D.H.; Park, K.-S.; Seo, S.-B. Regulation of UHRF1 acetylation by TIP60 is important for colon cancer cell proliferation. Genes Genom. 2022, 44, 1353–1361. [Google Scholar] [CrossRef]
- Tauber, M.; Fischle, W. Conserved linker regions and their regulation determine multiple chromatin-binding modes of UHRF1. Nucleus 2015, 6, 123–132. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Gao, Q.; Tan, S.; You, J.; Lyu, C.; Zhang, Y.; Han, M.; Chen, Z.; Li, J.; Wang, H.; et al. SET8 prevents excessive DNA methylation by methylation-mediated degradation of UHRF1 and DNMT1. Nucleic Acids Res. 2019, 47, 9053–9068. [Google Scholar] [CrossRef]
- Ding, G.; Chen, P.; Zhang, H.; Huang, X.; Zang, Y.; Li, J.; Li, J.; Wong, J. Regulation of Ubiquitin-like with Plant Homeodomain and RING Finger Domain 1 (UHRF1) Protein Stability by Heat Shock Protein 90 Chaperone Machinery. J. Biol. Chem. 2016, 291, 20125–20135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trotzier, M.-A.; Bronner, C.; Bathami, K.; Mathieu, E.; Abbady, A.-Q.; Jeanblanc, M.; Muller, C.D.; Rochette-Egly, C.; Mousli, M. Phosphorylation of ICBP90 by protein kinase A enhances topoisomerase IIα expression. Biochem. Biophys. Res. Commun. 2004, 319, 590–595. [Google Scholar] [CrossRef]
- Bronner, C.; Trotzier, M.-A.; Filhol, O.; Cochet, C.; Rochette-Egly, C.; Schöller-Guinard, M.; Klein, J.-P.; Mousli, M. The Antiapoptotic Protein ICBP90 Is a Target for Protein Kinase 2. Ann. N. Y. Acad. Sci. 2004, 1030, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, K.; Jin, B.; Chen, H.; Zhan, X.; Li, Z.; Wang, L.; Shen, X.; Li, M.; Yu, W.; et al. PIM1 induces cellular senescence through phosphorylation of UHRF1 at Ser311. Oncogene 2017, 36, 4828–4842. [Google Scholar] [CrossRef]
- Sidhu, H.; Capalash, N. UHRF1: The key regulator of epigenetics and molecular target for cancer therapeutics. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashraf, W.; Ibrahim, A.; Alhosin, M.; Zaayter, L.; Ouararhni, K.; Papin, C.; Ahmad, T.; Hamiche, A.; Mély, Y.; Bronner, C.; et al. The epigenetic integrator UHRF1: On the road to become a universal biomarker for cancer. Oncotarget 2017, 8, 51946–51962. [Google Scholar] [CrossRef] [Green Version]
- Abu-Alainin, W.; Gana, T.; Liloglou, T.; Olayanju, A.; Barrera, L.N.; Ferguson, R.; Campbell, F.; Andrews, T.; Goldring, C.; Kitteringham, N.; et al. UHRF1 regulation of the Keap1–Nrf2 pathway in pancreatic cancer contributes to oncogenesis. J. Pathol. 2016, 238, 423–433. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Shang, Y.; Jin, Z.; Zhang, W.; Lv, C.; Zhao, X.; Liu, Y.; Li, N.; Liang, J. UHRF1 promotes proliferation of gastric cancer via mediating tumor suppressor gene hypermethylation. Cancer Biol. Ther. 2015, 16, 1241–1251. [Google Scholar] [CrossRef] [Green Version]
- Daskalos, A.; Oleksiewicz, U.; Filia, A.; Nikolaidis, G.; Xinarianos, G.; Gosney, J.R.; Malliri, A.; Field, J.K.; Liloglou, T. UHRF1-mediated tumor suppressor gene inactivation in nonsmall cell lung cancer. Cancer 2011, 117, 1027–1037. [Google Scholar] [CrossRef]
- Mudbhary, R.; Hoshida, Y.; Chernyavskaya, Y.; Jacob, V.; Villanueva, A.; Fiel, M.I.; Chen, X.; Kojima, K.; Thung, S.; Bronson, R.T.; et al. UHRF1 Overexpression Drives DNA Hypomethylation and Hepatocellular Carcinoma. Cancer Cell 2014, 25, 196–209. [Google Scholar] [CrossRef] [Green Version]
- Babbio, F.; Pistore, C.; Curti, L.; Castiglioni, I.; Kunderfranco, P.; Brino, L.; Oudet, P.; Seiler, R.; Thalman, G.N.; Roggero, E.; et al. The SRA protein UHRF1 promotes epigenetic crosstalks and is involved in prostate cancer progression. Oncogene 2012, 31, 4878–4887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Huang, Z.; Zhu, Z.; Zheng, X.; Liu, J.; Han, Z.; Ma, X.; Zhang, Y. Upregulated UHRF1 Promotes Bladder Cancer Cell Invasion by Epigenetic Silencing of KiSS1. PLoS ONE 2014, 9, e104252. [Google Scholar] [CrossRef] [PubMed]
- Macaluso, M.; Montanari, M.; Noto, P.B.; Gregorio, V.; Bronner, C.; Giordano, A. Epigenetic Modulation of Estrogen Receptor-α by pRb Family Proteins: A Novel Mechanism in Breast Cancer. Cancer Res 2007, 67, 7731–7737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, W.; Chen, L.; Chen, Y.; Xu, S.-G.; Di, G.-H.; Yin, W.-J.; Wu, J.; Shao, Z.-M. UHRF1 is associated with epigenetic silencing of BRCA1 in sporadic breast cancer. Breast Cancer Res. Treat. 2009, 123, 359–373. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.-P.; Sun, H.-F.; Li, L.-D.; Fu, W.-Y.; Jin, W. UHRF1 Promotes Breast Cancer Progression by Suppressing KLF17 Ex-pression by Hypermethylating Its Promoter. Am. J. Cancer Res. 2017, 7, 1554. [Google Scholar]
- Beck, A.; Trippel, F.; Wagner, A.; Joppien, S.; Felle, M.; Vokuhl, C.; Schwarzmayr, T.; Strom, T.M.; von Schweinitz, D.; Längst, G.; et al. Overexpression of UHRF1 promotes silencing of tumor suppressor genes and predicts outcome in hepatoblastoma. Clin. Epigenetics 2018, 10, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdullah, O.; Omran, Z.; Hosawi, S.; Hamiche, A.; Bronner, C.; Alhosin, M. Thymoquinone Is a Multitarget Single Epidrug That Inhibits the UHRF1 Protein Complex. Genes 2021, 12, 622. [Google Scholar] [CrossRef]
- Nakamura, K.; Baba, Y.; Kosumi, K.; Harada, K.; Shigaki, H.; Miyake, K.; Kiyozumi, Y.; Ohuchi, M.; Kurashige, J.; Ishimoto, T.; et al. UHRF1 regulates global DNA hypomethylation and is associated with poor prognosis in esophageal squamous cell carcinoma. Oncotarget 2016, 7, 57821–57831. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.H.; Jin, E.; Kim, S.; Song, K.; Sung, J.K. LINE-1 hypomethylation is inversely correlated with UHRF1 overexpression in gastric cancer. Oncol. Lett. 2018, 15, 6666–6670. [Google Scholar] [CrossRef] [Green Version]
- Yuan, B.; Liu, Y.; Yu, X.; Yin, L.; Peng, Y.; Gao, Y.; Zhu, Q.; Cao, T.; Yang, Y.; Fan, X.; et al. FOXM1 contributes to taxane resistance by regulating UHRF1-controlled cancer cell stemness. Cell Death Dis. 2018, 9, 562. [Google Scholar] [CrossRef]
- Fu, Y.; Cao, T.; Zou, X.; Ye, Y.; Liu, Y.; Peng, Y.; Deng, T.; Yin, L.; Li, X. AKT1 regulates UHRF1 protein stability and promotes the resistance to abiraterone in prostate cancer. Oncogenesis 2023, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liu, Y.; Liu, Y.; Peng, Y.; Yuan, B.; Fu, Y.; Qi, X.; Zhu, Q.; Cao, T.; Zhang, S.; et al. UHRF1 promotes androgen receptor-regulated CDC6 transcription and anti-androgen receptor drug resistance in prostate cancer through KDM4C-Mediated chromatin modifications. Cancer Lett. 2021, 520, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Meng, Q.; Rosen, E.M.; Fan, S. UHRF1 confers radioresistance to human breast cancer cells. Int. J. Radiat. Biol. 2011, 87, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Liu, Y.; Peng, Y.; Peng, Y.; Yu, X.; Gao, Y.; Yuan, B.; Zhu, Q.; Cao, T.; He, L.; et al. Correction to: PARP inhibitor veliparib and HDAC inhibitor SAHA synergistically co-target the UHRF1/. BRCA1 DNA damage repair complex in prostate cancer cells. J. Exp. Clin. Cancer Res. 2022, 41, 1–2. [Google Scholar] [CrossRef]
- Liu, W.H.; Miner, R.E.; Albaugh, B.N.; Ananiev, G.E.; Wildman, S.A.; Denu, J.M. Discovery and Mechanism of Small Molecule Inhibitors Selective for the Chromatin-Binding Domains of Oncogenic UHRF1. Biochemistry 2022, 61, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Lou, C.; Ye, X.; Chen, G.; Zhu, J.; Kang, J. Screening inhibitors for blocking UHRF1-methylated DNA interaction with capillary electrophoresis. J. Chromatogr. A 2021, 1636, 461790. [Google Scholar] [CrossRef] [PubMed]
- Zaayter, L.; Mori, M.; Ahmad, T.; Ashraf, W.; Boudier, C.; Kilin, V.; Gavvala, K.; Richert, L.; Eiler, S.; Ruff, M.; et al. A Molecular Tool Targeting the Base-Flipping Activity of Human UHRF1. Chem. Eur. J. 2019, 25, 13363–13375. [Google Scholar] [CrossRef]
- Vaughan, R.M.; Dickson, B.M.; Whelihan, M.F.; Johnstone, A.L.; Cornett, E.M.; Cheek, M.A.; Ausherman, C.A.; Cowles, M.W.; Sun, Z.-W.; Rothbart, S.B. Chromatin structure and its chemical modifications regulate the ubiquitin ligase substrate selectivity of UHRF1. Proc. Natl. Acad. Sci. USA 2018, 115, 8775–8780. [Google Scholar] [CrossRef] [Green Version]
- Myrianthopoulos, V.; Cartron, P.F.; Liutkevičiūtė, Z.; Klimašauskas, S.; Matulis, D.; Bronner, C.; Martinet, N.; Mikros, E. Tandem virtual screening targeting the SRA domain of UHRF1 identifies a novel chemical tool modulating DNA methylation. Eur. J. Med. Chem. 2016, 114, 390–396. [Google Scholar] [CrossRef]
- Kilin, V.; Gavvala, K.; Barthes, N.P.F.; Michel, B.Y.; Shin, D.; Boudier, C.; Mauffret, O.; Yashchuk, V.; Mousli, M.; Ruff, M.; et al. Dynamics of Methylated Cytosine Flipping by UHRF1. J. Am. Chem. Soc. 2017, 139, 2520–2528. [Google Scholar] [CrossRef] [Green Version]
- Ciaco, S.; Mazzoleni, V.; Javed, A.; Eiler, S.; Ruff, M.; Mousli, M.; Mori, M.; Mély, Y. Inhibitors of UHRF1 base flipping activity showing cytotoxicity against cancer cells. Bioorgan. Chem. 2023, 137, 106616. [Google Scholar] [CrossRef] [PubMed]
- Giovinazzo, H.; Walker, D.; Wyhs, N.; Liu, J.; Esopi, D.M.; Vaghasia, A.M.; Jain, Y.; Bhamidipati, A.; Zhou, J.; Nelson, W.G. A High-Throughput Screen of Pharmacologically Active Compounds for Inhibitors of UHRF1 Reveals Epigenetic Activity of An-thracycline Derivative Chemotherapeutic Drugs. Oncotarget 2019, 10, 3040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houliston, R.S.; Lemak, A.; Iqbal, A.; Ivanochko, D.; Duan, S.; Kaustov, L.; Ong, M.S.; Fan, L.; Senisterra, G.; Brown, P.J.; et al. Conformational dynamics of the TTD–PHD histone reader module of the UHRF1 epigenetic regulator reveals multiple histone-binding states, allosteric regulation, and druggability. J. Biol. Chem. 2017, 292, 20947–20959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senisterra, G.; Zhu, H.Y.; Luo, X.; Zhang, H.; Xun, G.; Lu, C.; Xiao, W.; Hajian, T.; Loppnau, P.; Chau, I.; et al. Discovery of Small-Molecule Antagonists of the H3K9me3 Binding to UHRF1 Tandem Tudor Domain. SLAS Discov. Adv. Sci. Drug Discov. 2018, 23, 930–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kori, S.; Shibahashi, Y.; Ekimoto, T.; Nishiyama, A.; Yoshimi, S.; Yamaguchi, K.; Nagatoishi, S.; Ohta, M.; Tsumoto, K.; Nakanishi, M.; et al. Structure-based screening combined with computational and biochemical analyses identified the inhibitor targeting the binding of DNA Ligase 1 to UHRF1. Bioorgan. Med. Chem. 2021, 52, 116500. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-Y.; Cheng, Z.; Malbon, C.C. Overexpression of mitogen-activated protein kinase phosphatases MKP1, MKP2 in human breast cancer. Cancer Lett. 2003, 191, 229–237. [Google Scholar] [CrossRef]
- Huynh, H.; Soo, K.C.; Chow, P.K.; Tran, E. Targeted inhibition of the extracellular signal-regulated kinase kinase pathway with AZD6244 (ARRY-142886) in the treatment of hepatocellular carcinoma. Mol. Cancer Ther. 2007, 6, 138–146. [Google Scholar] [CrossRef] [Green Version]
- Fang, Z.; Xing, F.; Bronner, C.; Teng, Z.; Guo, Z. ICBP90 mediates the ERK1/2 signaling to regulate the proliferation of Jurkat T cells. Cell. Immunol. 2009, 257, 80–87. [Google Scholar] [CrossRef]
- Li, J.; Wang, R.; Hu, X.; Gao, Y.; Wang, Z.; Li, J.; Wong, J. Activated MEK/ERK Pathway Drives Widespread and Coordinated Overexpression of UHRF1 and DNMT1 in Cancer cells. Sci. Rep. 2019, 9, 907. [Google Scholar] [CrossRef] [Green Version]
- Morrissey, C.; Gallis, B.; Solazzi, J.W.; Kim, B.J.; Gulati, R.; Vakar-Lopez, F.; Goodlett, D.R.; Vessella, R.L.; Sasaki, T. Effect of artemisinin derivatives on apoptosis and cell cycle in prostate cancer cells. Anti-Cancer Drugs 2010, 21, 423–432. [Google Scholar] [CrossRef]
- Xu, C.-C.; Deng, T.; Fan, M.-L.; Lv, W.-B.; Liu, J.-H.; Yu, B.-Y. Synthesis and in vitro antitumor evaluation of dihydroartemisinin-cinnamic acid ester derivatives. Eur. J. Med. Chem. 2016, 107, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Xu, G.; Zou, W.; Xiang, T.; Luo, Z. Effect of dihydroartemisinin on UHRF1 gene expression in human prostate cancer PC-3 cells. Anti-Cancer Drugs 2017, 28, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Liu, S.; Xu, G.; Zhou, S.; Luo, Z. Dihydroartemisinin induces cell apoptosis through repression of UHRF1 in prostate cancer cells. Anti-Cancer Drugs 2021, 33, e113–e124. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, Q.; Fu, Y.; Ding, R.-B.; Qi, X.; Zhou, X.; Sun, Z.; Bao, J. Magnolol as a Potential Anticancer Agent: A Proposed Mechanistic Insight. Molecules 2022, 27, 6441. [Google Scholar] [CrossRef] [PubMed]
- Abid, R.; Ghazanfar, S.; Farid, A.; Sulaman, S.M.; Idrees, M.; Amen, R.A.; Muzammal, M.; Shahzad, M.K.; Mohamed, M.O.; Khaled, A.A.; et al. Pharmacological Properties of 4′, 5, 7-Trihydroxyflavone (Apigenin) and Its Impact on Cell Signaling Pathways. Molecules 2022, 27, 4304. [Google Scholar] [CrossRef] [PubMed]
- Luqman, S.; Meena, A.; Singh, J. Carvacrol as a prospective regulator of cancer targets/signalling pathways. Curr. Mol. Pharmacol. 2023, 16, 542–558. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, X.; Su, Z.; Fei, H.; Liu, X.; Pan, Q. The novel mTOR inhibitor Torin-2 induces autophagy and downregulates the expression of UHRF1 to suppress hepatocarcinoma cell growth. Oncol. Rep. 2015, 34, 1708–1716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Ou, H.; Xiang, L.; Li, X.; Huang, Y.; Yang, D. Elevated UHRF1 expression contributes to poor prognosis by promoting cell proliferation and metastasis in hepatocellular carcinoma. Oncotarget 2017, 8, 10510–10522. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Chen, B.; Zeng, Y.; Wang, H. UHRF1 Could Be a Prognostic Biomarker and Correlated with Immune Cell Infiltration in Hepatocellular Carcinoma. Int. J. Gen. Med. 2021, 14, 6769. [Google Scholar] [CrossRef]
- Awasthi, A.; Kumar, P.; Srikanth, C.V.; Sahi, S.; Puria, R. Invitro Evaluation of Torin2 and 2, 6-Dihydroxyacetophenone in Colorectal Cancer Therapy. Pathol. Oncol. Res. 2019, 25, 1121. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, J.; Liu, Q.; Ke, M.; Li, J.; Suo, W.; Guo, W.; Ma, A. Torin2 inhibits the EGFR-TKI resistant Non-Small Lung Cancer cell proliferation through negative feedback regulation of Akt/mTOR signaling. J. Cancer 2020, 11, 5746–5757. [Google Scholar] [CrossRef] [PubMed]
- Hui, B.; Pan, S.; Che, S.; Sun, Y.; Yan, Y.; Guo, J.; Gong, T.; Ren, J.; Zhang, X. Silencing UHRF1 Enhances Radiosensitivity of Esophageal Squamous Cell Carcinoma by Inhibiting the PI3K/Akt/mTOR Signaling Pathway. Cancer Manag. Res. 2021, 13, 4841–4852. [Google Scholar] [CrossRef] [PubMed]
- Cragg, G.M.; Newman, D.J. Plants as a source of anti-cancer agents. J. Ethnopharmacol. 2005, 100, 72–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, V.; Negi, A.S.; Kumar, J.; Gupta, M.; Khanuja, S.P. Plant-based anticancer molecules: A chemical and biological profile of some important leads. Bioorgan. Med. Chem. 2005, 13, 5892–5908. [Google Scholar] [CrossRef] [PubMed]
- Awal, A.; Nur, S.M.; Al Khalaf, A.K.; Rehan, M.; Ahmad, A.; Hosawi, S.B.I.; Choudhry, H.; Khan, M.I. Structural-Guided Identification of Small Molecule Inhibitor of UHRF1 Methyltransferase Activity. Front. Genet. 2022, 13, 928884. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Yin, Z.; Nie, H.; Liu, Y.; Yang, J.; Huang, G.; Shen, J.; Chen, L.; Fei, J. Identification of berberine as a novel drug for the treatment of multiple myeloma via targeting UHRF1. BMC Biol. 2020, 18, 33. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Campbell, J.; Raji, I.O.; Guduru, S.K.R.; Kandel, P.; Nguyen, M.; Liu, S.; Tran, K.; Venugopal, N.K.; Taylor, B.C.; et al. Discovery of small molecules targeting the tandem tudor domain of the epigenetic factor UHRF1 using fragment-based ligand discovery. Sci. Rep. 2021, 11, 1–17. [Google Scholar] [CrossRef]
- Patnaik, D.; Estève, P.-O.; Pradhan, S. Targeting the SET and RING-associated (SRA) domain of ubiquitin-like, PHD and ring finger-containing 1 (UHRF1) for anti-cancer drug development. Oncotarget 2018, 9, 26243–26258. [Google Scholar] [CrossRef] [Green Version]
- Skała, E.; Toma, M.; Kowalczyk, T.; Śliwiński, T.; Sitarek, P. Rhaponticum carthamoides transformed root extract inhibits human glioma cells viability, induces double strand DNA damage, H2A.X phosphorylation, and PARP1 cleavage. Cytotechnology 2018, 70, 1585–1594. [Google Scholar] [CrossRef] [Green Version]
- Sitarek, P.; Kowalczyk, T.; Santangelo, S.; Bialas, A.; Toma, M.; Wieczfinska, J.; Śliwiński, T.; Skała, E. The Extract of Leonurus sibiricus Transgenic Roots with AtPAP1 Transcriptional Factor Induces Apoptosis via DNA Damage and Down Regulation of Selected Epigenetic Factors in Human Cancer Cells. Neurochem. Res. 2018, 43, 1363–1370. [Google Scholar] [CrossRef] [Green Version]
- Mauray, A.; Milenkovic, D.; Besson, C.; Caccia, N.; Morand, C.; Michel, F.; Mazur, A.; Scalbert, A.; Felgines, C. Atheroprotective Effects of Bilberry Extracts in Apo E-Deficient Mice. J. Agric. Food Chem. 2009, 57, 11106–11111. [Google Scholar] [CrossRef] [PubMed]
- Walter, A.; Etienne-Selloum, N.; Brasse, D.; Khallouf, H.; Bronner, C.; Rio, M.; Beretz, A.; Schini-Kerth, V.B. Intake of Grape-derived Polyphenols Reduces C26 Tumor Growth by Inhibiting Angiogenesis and Inducing Apoptosis. FASEB J. 2010, 24, 3360–3369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alhosin, M.; León-González, A.J.; Dandache, I.; Lelay, A.; Rashid, S.K.; Kevers, C.; Pincemail, J.; Fornecker, L.-M.; Mauvieux, L.; Herbrecht, R.; et al. Bilberry extract (Antho 50) selectively induces redox-sensitive caspase 3-related apoptosis in chronic lymphocytic leukemia cells by targeting the Bcl-2/Bad pathway. Sci. Rep. 2015, 5, 8996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- León-González, A.J.; Sharif, T.; Auger, C.; Abbas, M.; Fuhrmann, G.; Schini-Kerth, V.B. Anthocyanin-rich bilberry extract induces apoptosis in acute lymphoblastic leukemia cells via redox-sensitive epigenetic modifications. J. Funct. Foods 2018, 44, 227–234. [Google Scholar] [CrossRef]
- Sharif, T.; Alhosin, M.; Auger, C.; Minker, C.; Kim, J.-H.; Etienne-Selloum, N.; Bories, P.; Gronemeyer, H.; Lobstein, A.; Bronner, C.; et al. Aronia melanocarpa Juice Induces a Redox-Sensitive p73-Related Caspase 3-Dependent Apoptosis in Human Leukemia Cells. PLoS ONE 2012, 7, e32526. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, W.; Ahmad, T.; Almalki, N.A.R.; Krifa, M.; Zaayter, L.; Pizzi, A.; Muller, C.D.; Hamiche, A.; Mély, Y.; Bronner, C.; et al. Tannin extract from maritime pine bark exhibits anticancer properties by targeting the epigenetic UHRF1/DNMT1 tandem leading to the re-expression of TP73. Food Funct. 2022, 13, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Sharif, T.; Auger, C.; Alhosin, M.; Ebel, C.; Achour, M.; Étienne-Selloum, N.; Fuhrmann, G.; Bronner, C.; Schini-Kerth, V.B. Red wine polyphenols cause growth inhibition and apoptosis in acute lymphoblastic leukaemia cells by inducing a redox-sensitive up-regulation of p73 and down-regulation of UHRF1. Eur. J. Cancer 2010, 46, 983–994. [Google Scholar] [CrossRef] [PubMed]
- Parashar, G.; Capalash, N. Promoter methylation-independent reactivation of PAX1 by curcumin and resveratrol is mediated by UHRF1. Clin. Exp. Med. 2016, 16, 471–478. [Google Scholar] [CrossRef]
- Krifa, M.; Alhosin, M.; Muller, C.D.; Gies, J.-P.; Chekir-Ghedira, L.; Ghedira, K.; Mély, Y.; Bronner, C.; Mousli, M. Limoniastrum guyonianum aqueous gall extract induces apoptosis in human cervical cancer cells involving p16INK4A re-expression related to UHRF1 and DNMT1 down-regulation. CR J. Exp. Clin. Cancer Res. 2013, 32, 30. [Google Scholar] [CrossRef] [Green Version]
- Hasan, M.N.; Choudhry, H.; Razvi, S.S.; Moselhy, S.S.; Kumosani, T.A.; Zamzami, M.A.; Omran, Z.; Halwani, M.A.; Al-Babili, S.; Abualnaja, K.O.; et al. Synthetic strigolactone analogues reveal anti-cancer activities on hepatocellular carcinoma cells. Bioorgan. Med. Chem. Lett. 2018, 28, 1077–1083. [Google Scholar] [CrossRef] [Green Version]
- Hasan, M.N.; Razvi, S.S.; Choudhry, H.; Hassan, M.A.; Moselhy, S.S.; Kumosani, T.A.; Zamzami, M.A.; Abualnaja, K.O.; Halwani, M.A.; Al-Malki, A.L.; et al. Gene Ontology and Expression Studies of Strigolactone Analogues on a Hepatocellular Carcinoma Cell Line. Anal. Cell. Pathol. 2019, 2019, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krifa, M.; Leloup, L.; Ghedira, K.; Mousli, M.; Chekir-Ghedira, L. Luteolin Induces Apoptosis in BE Colorectal Cancer Cells by Downregulating Calpain, UHRF1, and DNMT1 Expressions. Nutr. Cancer 2014, 66, 1220–1227. [Google Scholar] [CrossRef] [PubMed]
- Achour, M.; Mousli, M.; Alhosin, M.; Ibrahim, A.; Peluso, J.; Muller, C.D.; Schini-Kerth, V.B.; Hamiche, A.; Dhe-Paganon, S.; Bronner, C. Epigallocatechin-3-gallate up-regulates tumor suppressor gene expression via a reactive oxygen species-dependent down-regulation of UHRF1. Biochem. Biophys. Res. Commun. 2013, 430, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Pozo, F.M.; Tian, D.; Geng, X.; Yao, X.; Zhang, Y.; Tang, J. Shikonin Inhibits Cancer Through P21 Upregulation and Apoptosis Induction. Front. Pharmacol. 2020, 11, 861. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.Y.; Hong, D.; Jeong, S.Y.; Kim, J.-H. Shikonin causes apoptosis by up-regulating p73 and down-regulating ICBP90 in human cancer cells. Biochem. Biophys. Res. Commun. 2015, 465, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wu, C.; Muhammad, J.S.; Yan, D.; Tsuneyama, K.; Hatta, H.; Cui, Z.-G.; Inadera, H. Melatonin sensitises shikonin-induced cancer cell death mediated by oxidative stress via inhibition of the SIRT3/SOD2-AKT pathway. Redox Biol. 2020, 36, 101632. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.S.; Choi, Y.H.; Moon, J.W.; Kim, H.S.; Park, S.-H. Hinokitiol induces DNA demethylation via DNMT1 and UHRF1 inhibition in colon cancer cells. BMC Cell Biol. 2017, 18, 14. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Chen, W.; Wang, Z.; Cai, P. Emodin promotes the arrest of human lymphoma Raji cell proliferation through the UHRF1-DNMT3A-∆Np73 pathways. Mol. Med. Rep. 2017, 16, 6544–6551. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Tang, R.; Ding, L.; Zheng, R.; Liu, Y.; Yin, L.; Fu, Y.; Deng, T.; Li, X. Diosgenin inhibits prostate cancer progression by inducing UHRF1 protein degradation. Eur. J. Pharmacol. 2023, 942, 175522. [Google Scholar] [CrossRef]
- Lee, Y.-M.; Kim, G.-H.; Park, E.-J.; Oh, T.-I.; Lee, S.; Kan, S.-Y.; Kang, H.; Kim, B.M.; Kim, J.H.; Lim, J.-H. Thymoquinone Selectively Kills Hypoxic Renal Cancer Cells by Suppressing HIF-1α-Mediated Glycolysis. Int. J. Mol. Sci. 2019, 20, 1092. [Google Scholar] [CrossRef] [Green Version]
- Ndreshkjana, B.; Çapci, A.; Klein, V.; Chanvorachote, P.; Muenzner, J.K.; Huebner, K.; Steinmann, S.; Erlenbach-Wuensch, K.; Geppert, C.I.; Agaimy, A.; et al. Combination of 5-fluorouracil and thymoquinone targets stem cell gene signature in colorectal cancer cells. Cell Death Dis. 2019, 10, 379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Fan, Y.; Huang, S.; Wang, G.; Han, R.; Lei, F.; Luo, A.; Jing, X.; Zhao, L.; Gu, S.; et al. Thymoquinone inhibits the metastasis of renal cell cancer cells by inducing autophagy via AMPK/mTOR signaling pathway. Cancer Sci. 2018, 109, 3865–3873. [Google Scholar] [CrossRef] [PubMed]
- Darakhshan, S.; Bidmeshki Pour, A.; Hosseinzadeh Colagar, A.H.; Sisakhtnezhad, S. Thymoquinone and its therapeutic potentials. Pharmacol. Res. 2015, 95–96, 138–158. [Google Scholar] [CrossRef] [PubMed]
- Gali-Muhtasib, H.; Roessner, A.; Schneider-Stock, R. Thymoquinone: A promising anti-cancer drug from natural sources. Int. J. Biochem. Cell Biol. 2006, 38, 1249–1253. [Google Scholar] [CrossRef] [PubMed]
- Shoieb, A.M.; Elgayyar, M.; Dudrick, P.S.; Bell, J.L.; Tithof, P.K. In vitro inhibition of growth and induction of apoptosis in cancer cell lines by thymoquinone. Int. J. Oncol. 2003, 22, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Alhosin, M.; Abusnina, A.; Achour, M.; Sharif, T.; Muller, C.; Peluso, J.; Chataigneau, T.; Lugnier, C.; Schini-Kerth, V.B.; Bronner, C.; et al. Induction of apoptosis by thymoquinone in lymphoblastic leukemia Jurkat cells is mediated by a p73-dependent pathway which targets the epigenetic integrator UHRF1. Biochem. Pharmacol. 2010, 79, 1251–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibrahim, A.; Alhosin, M.; Papin, C.; Ouararhni, K.; Omran, Z.; Zamzami, M.A.; Al-Malki, A.L.; Choudhry, H.; Mély, Y.; Hamiche, A.; et al. Thymoquinone challenges UHRF1 to commit auto-ubiquitination: A key event for apoptosis induction in cancer cells. Oncotarget 2018, 9, 28599. [Google Scholar] [CrossRef] [Green Version]
- Polepalli, S.; George, S.M.; Vidya, R.V.S.; Rodrigues, G.S.; Ramachandra, L.; Chandrashekar, R.; Nayak, D.; Rao, P.P.; Pestell, R.G.; Rao, M. Role of UHRF1 in malignancy and its function as a therapeutic target for molecular docking towards the SRA domain. Int. J. Biochem. Cell Biol. 2019, 114, 105558. [Google Scholar] [CrossRef]
- Kim, M.Y.; Park, S.-J.; Shim, J.W.; Yang, K.; Kang, H.S.; Heo, K. Naphthazarin enhances ionizing radiation-induced cell cycle arrest and apoptosis in human breast cancer cells. Int. J. Oncol. 2015, 46, 1659–1666. [Google Scholar] [CrossRef] [Green Version]
- Luo, G.; Li, Q.; Yu, M.; Wang, T.; Zang, Y.; Liu, Z.; Niu, Z.; Yang, H.; Lai, J. UHRF1 modulates breast cancer cell growth via estrogen signaling. Med. Oncol. 2022, 39, 111. [Google Scholar] [CrossRef]
- Chow, M.; Gao, L.; MacManiman, J.D.; Bicocca, V.T.; Chang, B.H.; Alumkal, J.J.; Tyner, J.W. Maintenance and pharmacologic targeting of ROR1 protein levels via UHRF1 in t(1;19) pre-B-ALL. Oncogene 2018, 37, 5221–5232. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, H.; Capalash, N. Plumbagin downregulates UHRF1, p-Akt, MMP-2 and suppresses survival, growth and migration of cervical cancer CaSki cells. Toxicol. Vitr. 2023, 86, 105512. [Google Scholar] [CrossRef] [PubMed]
- Abusnina, A.; Keravis, T.; Yougbaré, I.; Bronner, C.; Lugnier, C. Anti-proliferative effect of curcumin on melanoma cells is mediated by PDE1A inhibition that regulates the epigenetic integrator UHRF1. Mol. Nutr. Food Res. 2011, 55, 1677–1689. [Google Scholar] [CrossRef] [PubMed]
- Majchrzak-Celińska, A.; Kleszcz, R.; Stasiłowicz-Krzemień, A.; Cielecka-Piontek, J. Sodium Butyrate Enhances Curcuminoids Permeability through the Blood-Brain Barrier, Restores Wnt/β-Catenin Pathway Antagonists Gene Expression and Reduces the Viability of Glioblastoma Cells. Int. J. Mol. Sci. 2021, 22, 11285. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Xing, F.; Tang, Z.; Bronner, C.; Lu, X.; Di, J.; Zeng, S.; Liu, J. Anisomycin suppresses Jurkat T cell growth by the cell cycle-regulating proteins. Pharmacol. Rep. 2013, 65, 435–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reardon, E.S.; Shukla, V.; Xi, S.; Gara, S.K.; Liu, Y.; Straughan, D.; Zhang, M.; Hong, J.A.; Payabyab, E.C.; Kumari, A.; et al. UHRF1 Is a Novel Druggable Epigenetic Target in Malignant Pleural Mesothelioma. J. Thorac. Oncol. 2021, 16, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Unoki, M.; Brunet, J.; Mousli, M. Drug discovery targeting epigenetic codes: The great potential of UHRF1, which links DNA methylation and histone modifications, as a drug target in cancers and toxoplasmosis. Biochem. Pharmacol. 2009, 78, 1279–1288. [Google Scholar] [CrossRef] [PubMed]
- Alhosin, M.; Omran, Z.; Zamzami, M.A.; Al-Malki, A.L.; Choudhry, H.; Mousli, M.; Bronner, C. Signalling pathways in UHRF1-dependent regulation of tumor suppressor genes in cancer. J. Exp. Clin. Cancer Res. 2016, 35, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Lyubitelev, A.; Studitsky, V. Inhibition of Cancer Development by Natural Plant Polyphenols: Molecular Mechanisms. Int. J. Mol. Sci. 2023, 24, 10663. [Google Scholar] [CrossRef]
- Kanwal, Q.; Ahmed, M.; Hamza, M.; Ahmad, M.; Rehman, A.U.; Yousaf, N.; Javaid, A.; Anwar, A.; Khan, I.H.; Muddassar, M. Curcumin nanoparticles: Physicochemical fabrication, characterization, antioxidant, enzyme inhibition, molecular docking and simulation studies. RSC Adv. 2023, 13, 22268–22280. [Google Scholar] [CrossRef]
- Coelho, S.C.; Estevinho, B.N. A Brief Review on the Electrohydrodynamic Techniques Used to Build Antioxidant Delivery Systems from Natural Sources. Molecules 2023, 28, 3592. [Google Scholar] [CrossRef]
- Mishra, A.; Gupta, M. Bioactive Flavonoids: A comparative overview of the biogenetic and chemical synthesis approach. Mini-Rev. Med. Chem. 2023, 23, 1–20. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashraf, W.; Ahmad, T.; Reynoird, N.; Hamiche, A.; Mély, Y.; Bronner, C.; Mousli, M. Natural and Synthetic Anticancer Epidrugs Targeting the Epigenetic Integrator UHRF1. Molecules 2023, 28, 5997. https://doi.org/10.3390/molecules28165997
Ashraf W, Ahmad T, Reynoird N, Hamiche A, Mély Y, Bronner C, Mousli M. Natural and Synthetic Anticancer Epidrugs Targeting the Epigenetic Integrator UHRF1. Molecules. 2023; 28(16):5997. https://doi.org/10.3390/molecules28165997
Chicago/Turabian StyleAshraf, Waseem, Tanveer Ahmad, Nicolas Reynoird, Ali Hamiche, Yves Mély, Christian Bronner, and Marc Mousli. 2023. "Natural and Synthetic Anticancer Epidrugs Targeting the Epigenetic Integrator UHRF1" Molecules 28, no. 16: 5997. https://doi.org/10.3390/molecules28165997
APA StyleAshraf, W., Ahmad, T., Reynoird, N., Hamiche, A., Mély, Y., Bronner, C., & Mousli, M. (2023). Natural and Synthetic Anticancer Epidrugs Targeting the Epigenetic Integrator UHRF1. Molecules, 28(16), 5997. https://doi.org/10.3390/molecules28165997