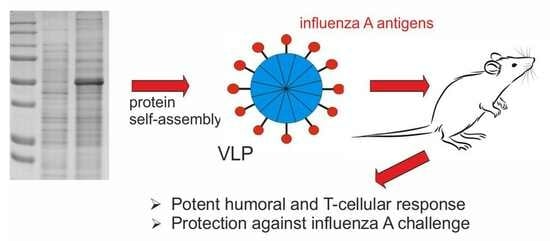
Nanoparticles Carrying Conserved Regions of Influenza A Hemagglutinin, Nucleoprotein, and M2 Protein Elicit a Strong Humoral and T Cell Immune Response and Protect Animals from Infection
Abstract
:1. Introduction
2. Results
2.1. Design of Recombinant Proteins
2.2. Expression and Purification of Recombinant Proteins
2.3. Formation of Nanoparticles
2.4. Humoral Immune Response
2.5. T Cellular Immune Response
2.6. Protection against Lethal Influenza Challenge
3. Discussion
4. Materials and Methods
4.1. Components of SAP-Based Fusion Proteins
4.2. Genes Encoding Fusion Proteins and Expression Vectors
4.3. Expression and Purification of Fusion Proteins
4.4. Western Blot Analysis of Protein Samples
4.5. Structural Analysis of Recombinant Proteins
4.6. Immunization of Animals
4.7. Antibody Detection in the Sera and BAL by ELISA
4.8. Isolation of Cells from the Spleen to Analyze the T Cellular Immune Response
4.9. Determination of Antigen-Specific Effector Memory T cells
4.10. Influenza Virus and Challenge Experiment
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Gerhard, W.; Mozdzanowska, K.; Zharikova, D. Prospects for Universal Influenza Virus Vaccine. Emerg. Infect. Dis. 2006, 12, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Palese, P.; García-Sastre, A. Influenza Vaccines: Present and Future. J. Clin. Investig. 2002, 110, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Saelens, X. The Role of Matrix Protein 2 Ectodomain in the Development of Universal Influenza Vaccines. J. Infect. Dis. 2019, 219, S68–S74. [Google Scholar] [CrossRef]
- Schotsaert, M.; De Filette, M.; Fiers, W.; Saelens, X. Universal M2 Ectodomain-Based Influenza A Vaccines: Preclinical and Clinical Developments. Expert Rev. Vaccines 2009, 8, 499–508. [Google Scholar] [CrossRef]
- Jegerlehner, A.; Schmitz, N.; Storni, T.; Bachmann, M.F. Influenza A Vaccine Based on the Extracellular Domain of M2: Weak Protection Mediated via Antibody-Dependent NK Cell Activity. J. Immunol. 2004, 172, 5598–5605. [Google Scholar] [CrossRef] [PubMed]
- El Bakkouri, K.; Descamps, F.; De Filette, M.; Smet, A.; Festjens, E.; Birkett, A.; Van Rooijen, N.; Verbeek, S.; Fiers, W.; Saelens, X. Universal Vaccine Based on Ectodomain of Matrix Protein 2 of Influenza A: Fc Receptors and Alveolar Macrophages Mediate Protection. J. Immunol. 2011, 186, 1022–1031. [Google Scholar] [CrossRef]
- Corti, D.; Voss, J.; Gamblin, S.J.; Codoni, G.; Macagno, A.; Jarrossay, D.; Vachieri, S.G.; Pinna, D.; Minola, A.; Vanzetta, F.; et al. A Neutralizing Antibody Selected from Plasma Cells That Binds to Group 1 and Group 2 Influenza A Hemagglutinins. Science 2011, 333, 850–856. [Google Scholar] [CrossRef] [PubMed]
- Tsybalova, L.M.; Stepanova, L.A.; Shuklina, M.A.; Mardanova, E.S.; Kotlyarov, R.Y.; Potapchuk, M.V.; Petrov, S.A.; Blokhina, E.A.; Ravin, N.V. Combination of M2e Peptide with Stalk HA Epitopes of Influenza A Virus Enhances Protective Properties of Recombinant Vaccine. PLoS ONE 2018, 13, e0201429. [Google Scholar] [CrossRef]
- Stepanova, L.A.; Mardanova, E.S.; Shuklina, M.A.; Blokhina, E.A.; Kotlyarov, R.Y.; Potapchuk, M.V.; Kovaleva, A.A.; Vidyaeva, I.G.; Korotkov, A.V.; Eletskaya, E.I.; et al. Flagellin-Fused Protein Targeting M2e and HA2 Induces Potent Humoral and T-Cell Responses and Protects Mice against Various Influenza Viruses a Subtypes. J. Biomed. Sci. 2018, 25, 33. [Google Scholar] [CrossRef]
- Jegaskanda, S.; Job, E.R.; Kramski, M.; Laurie, K.; Isitman, G.; de Rose, R.; Winnall, W.R.; Stratov, I.; Brooks, A.G.; Reading, P.C.; et al. Cross-Reactive Influenza-Specific Antibody-Dependent Cellular Cytotoxicity Antibodies in the Absence of Neutralizing Antibodies. J. Immunol. 2013, 190, 1837–1848. [Google Scholar] [CrossRef]
- DiLillo, D.J.; Tan, G.S.; Palese, P.; Ravetch, J.V. Broadly Neutralizing Hemagglutinin Stalk-Specific Antibodies Require FcγR Interactions for Protection against Influenza Virus in Vivo. Nat. Med. 2014, 20, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Epstein, S.L. Universal Influenza Vaccines: Progress in Achieving Broad Cross-Protection In Vivo. Am. J. Epidemiol. 2018, 187, 2603–2614. [Google Scholar] [CrossRef] [PubMed]
- Shu, L.L.; Bean, W.J.; Webster, R.G. Analysis of the Evolution and Variation of the Human Influenza A Virus Nucleoprotein Gene from 1933 to 1990. J. Virol. 1993, 67, 2723–2729. [Google Scholar] [CrossRef]
- Yewdell, J.W.; Bennink, J.R.; Smith, G.L.; Moss, B. Influenza A Virus Nucleoprotein Is a Major Target Antigen for Cross-Reactive Anti-Influenza A Virus Cytotoxic T Lymphocytes. Proc. Natl. Acad. Sci. USA 1985, 82, 1785–1789. [Google Scholar] [CrossRef]
- Townsend, A.R.M.; Gotch, F.M.; Davey, J. Cytotoxic T Cells Recognize Fragments of the Influenza Nucleoprotein. Cell 1985, 42, 457–467. [Google Scholar] [CrossRef]
- Brown, L.E.; Kelso, A. Prospects for an Influenza Vaccine That Induces Cross-protective Cytotoxic T Lymphocytes. Immunol. Cell. Biol. 2009, 87, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Romeli, S.; Hassan, S.S.; Yap, W.B. Multi-Epitope Peptide-Based and Vaccinia-Based Universal Influenza Vaccine Candidates Subjected to Clinical Trials. MJMS 2020, 27, 10–20. [Google Scholar] [CrossRef]
- Van Doorn, E.; Liu, H.; Ben-Yedidia, T.; Hassin, S.; Visontai, I.; Norley, S.; Frijlink, H.W.; Hak, E. Evaluating the Immunogenicity and Safety of a BiondVax-Developed Universal Influenza Vaccine (Multimeric-001) Either as a Standalone Vaccine or as a Primer to H5N1 Influenza Vaccine: Phase IIb Study Protocol. Medicine 2017, 96, e6339. [Google Scholar] [CrossRef]
- Pleguezuelos, O.; Robinson, S.; Stoloff, G.A.; Caparrós-Wanderley, W. Synthetic Influenza Vaccine (FLU-v) Stimulates Cell Mediated Immunity in a Double-Blind, Randomised, Placebo-Controlled Phase I Trial. Vaccine 2012, 30, 4655–4660. [Google Scholar] [CrossRef]
- Burkhard, P.; Meier, M.; Lustig, A. Design of a Minimal Protein Oligomerization Domain by a Structural Approach. Protein Sci. 2000, 9, 2294–2301. [Google Scholar] [CrossRef]
- Doll, T.A.P.F.; Dey, R.; Burkhard, P. Design and Optimization of Peptide Nanoparticles. J. Nanobiotechnol. 2015, 13, 73. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, T.A.P.F.; Yan, Z.; Jeffers, S.A.; Holmes, K.V.; Hodges, R.S.; Burkhard, P. Peptide Nanoparticles as Novel Immunogens: Design and Analysis of a Prototypic Severe Acute Respiratory Syndrome Vaccine. Chem. Biol. Drug Des. 2009, 73, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Kaba, S.A.; Brando, C.; Guo, Q.; Mittelholzer, C.; Raman, S.; Tropel, D.; Aebi, U.; Burkhard, P.; Lanar, D.E. A Nonadjuvanted Polypeptide Nanoparticle Vaccine Confers Long-Lasting Protection against Rodent Malaria. J. Immunol. 2009, 183, 7268–7277. [Google Scholar] [CrossRef] [PubMed]
- Wahome, N.; Pfeiffer, T.; Ambiel, I.; Yang, Y.; Keppler, O.T.; Bosch, V.; Burkhard, P. Conformation-Specific Display of 4E10 and 2F5 Epitopes on Self-Assembling Protein Nanoparticles as a Potential HIV Vaccine: MPER-Specific Protein Nanoparticle HIV Vaccine. Chem. Biol. Drug Des. 2012, 80, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Babapoor, S.; Neef, T.; Mittelholzer, C.; Girshick, T.; Garmendia, A.; Shang, H.; Khan, M.I.; Burkhard, P. A Novel Vaccine Using Nanoparticle Platform to Present Immunogenic M2e against Avian Influenza Infection. Influenza Res. Treat. 2011, 2011, 126794. [Google Scholar] [CrossRef]
- Raman, S.; Machaidze, G.; Lustig, A.; Aebi, U.; Burkhard, P. Structure-Based Design of Peptides That Self-Assemble into Regular Polyhedral Nanoparticles. Nanomedicine 2006, 2, 95–102. [Google Scholar] [CrossRef]
- Zykova, A.A.; Blokhina, E.A.; Stepanova, L.A.; Shuklina, M.A.; Tsybalova, L.M.; Kuprianov, V.V.; Ravin, N.V. Nanoparticles Based on Artificial Self-Assembling Peptide and Displaying M2e Peptide and Stalk HA Epitopes of Influenza A Virus Induce Potent Humoral and T-Cell Responses and Protect against the Viral Infection. Nanomedicine 2022, 39, 102463. [Google Scholar] [CrossRef]
- Alexander, J.; Del Guercio, M.-F.; Maewal, A.; Qiao, L.; Fikes, J.; Chesnut, R.W.; Paulson, J.; Bundle, D.R.; DeFrees, S.; Sette, A. Linear PADRE T Helper Epitope and Carbohydrate B Cell Epitope Conjugates Induce Specific High Titer IgG Antibody Responses. J. Immunol. 2000, 164, 1625–1633. [Google Scholar] [CrossRef]
- Ravin, N.V.; Blokhina, E.A.; Kuprianov, V.V.; Stepanova, L.A.; Shaldjan, A.A.; Kovaleva, A.A.; Tsybalova, L.M.; Skryabin, K.G. Development of a Candidate Influenza Vaccine Based on Virus-like Particles Displaying Influenza M2e Peptide into the Immunodominant Loop Region of Hepatitis B Core Antigen: Insertion of Multiple Copies of M2e Increases Immunogenicity and Protective Efficiency. Vaccine 2015, 33, 3392–3397. [Google Scholar] [CrossRef]
- Jennings, G.T.; Bachmann, M.F. The Coming of Age of Virus-like Particle Vaccines. Biol. Chem. 2008, 389, 521–536. [Google Scholar] [CrossRef]
- Al-Halifa, S.; Gauthier, L.; Arpin, D.; Bourgault, S.; Archambault, D. Nanoparticle-Based Vaccines Against Respiratory Viruses. Front. Immunol. 2019, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Huber, V.C.; Lynch, J.M.; Bucher, D.J.; Le, J.; Metzger, D.W. Fc Receptor-Mediated Phagocytosis Makes a Significant Contribution to Clearance of Influenza Virus Infections. J. Immunol. 2001, 166, 7381–7388. [Google Scholar] [CrossRef]
- Jegaskanda, S.; Vanderven, H.A.; Wheatley, A.K.; Kent, S.J. Fc or Not Fc; That Is the Question: Antibody Fc-Receptor Interactions Are Key to Universal Influenza Vaccine Design. Hum. Vaccin. Immunother. 2017, 13, 1288–1296. [Google Scholar] [CrossRef] [PubMed]
- Wohlbold, T.J.; Chromikova, V.; Tan, G.S.; Meade, P.; Amanat, F.; Comella, P.; Hirsh, A.; Krammer, F. Hemagglutinin Stalk- and Neuraminidase-Specific Monoclonal Antibodies Protect against Lethal H10N8 Influenza Virus Infection in Mice. J. Virol. 2016, 90, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-Q.; Lan, L.Y.-L.; Huang, M.; Henry, C.; Wilson, P.C. Hemagglutinin Stalk-Reactive Antibodies Interfere with Influenza Virus Neuraminidase Activity by Steric Hindrance. J. Virol. 2019, 93, e01526-18. [Google Scholar] [CrossRef] [PubMed]
- Bender, B.S.; Croghan, T.; Zhang, L.; Small, P.A. Transgenic Mice Lacking Class I Major Histocompatibility Complex-Restricted T Cells Have Delayed Viral Clearance and Increased Mortality after Influenza Virus Challenge. J. Exp. Med. 1992, 175, 1143–1145. [Google Scholar] [CrossRef] [PubMed]
- Doherty, P.C.; Topham, D.J.; Tripp, R.A.; Cardin, R.D.; Brooks, J.W.; Stevenson, P.G. Effector CD4+ and CD8+ T-Cell Mechanisms in the Control of Respiratory Virus Infections. Immunol. Rev. 1997, 159, 105–117. [Google Scholar] [CrossRef]
- Gerhard, W. The Role of the Antibody Response in Influenza Virus Infection. In Antibodies in Viral Infection; Current Topics in Microbiology and Immunology; Burton, D.R., Ed.; Springer: Berlin/Heidelberg, Germany, 2001; Volume 260, pp. 171–190. ISBN 978-3-642-07486-8. [Google Scholar]
- Thomas, P.G.; Keating, R.; Hulse-Post, D.J.; Doherty, P.C. Cell-Mediated Protection in Influenza Infection. Emerg. Infect. Dis. 2006, 12, 48–54. [Google Scholar] [CrossRef]
- Chalifour, A.; Jeannin, P.; Gauchat, J.-F.; Blaecke, A.; Malissard, M.; N’Guyen, T.; Thieblemont, N.; Delneste, Y. Direct Bacterial Protein PAMP Recognition by Human NK Cells Involves TLRs and Triggers α-Defensin Production. Blood 2004, 104, 1778–1783. [Google Scholar] [CrossRef]
- Kannan, K.; Stewart, R.M.; Bounds, W.; Carlsson, S.R.; Fukuda, M.; Betzing, K.W.; Holcombe, R.F. Lysosome-Associated Membrane Proteins h-LAMP1 (CD107a) and h-LAMP2 (CD107b) Are Activation-Dependent Cell Surface Glycoproteins in Human Peripheral Blood Mononuclear Cells Which Mediate Cell Adhesion to Vascular Endothelium. Cell. Immunol. 1996, 171, 10–19. [Google Scholar] [CrossRef]
- Clemens, E.; Van De Sandt, C.; Wong, S.; Wakim, L.; Valkenburg, S. Harnessing the Power of T Cells: The Promising Hope for a Universal Influenza Vaccine. Vaccines 2018, 6, 18. [Google Scholar] [CrossRef]
- Sridhar, S. Heterosubtypic T-Cell Immunity to Influenza in Humans: Challenges for Universal T-Cell Influenza Vaccines. Front. Immunol. 2016, 7, 195. [Google Scholar] [CrossRef]
- De Filette, M.; Min Jou, W.; Birkett, A.; Lyons, K.; Schultz, B.; Tonkyro, A.; Resch, S.; Fiers, W. Universal Influenza A Vaccine: Optimization of M2-Based Constructs. Virology 2005, 337, 149–161. [Google Scholar] [CrossRef]
- Stoloff, G.A.; Caparros-Wanderley, W. Synthetic Multi-Epitope Peptides Identified in Silico Induce Protective Immunity against Multiple Influenza Serotypes. Eur. J. Immunol. 2007, 37, 2441–2449. [Google Scholar] [CrossRef]
- Atsmon, J.; Kate-Ilovitz, E.; Shaikevich, D.; Singer, Y.; Volokhov, I.; Haim, K.Y.; Ben-Yedidia, T. Safety and Immunogenicity of Multimeric-001—A Novel Universal Influenza Vaccine. J. Clin. Immunol. 2012, 32, 595–603. [Google Scholar] [CrossRef]
- Arai, R.; Ueda, H.; Kitayama, A.; Kamiya, N.; Nagamune, T. Design of the Linkers Which Effectively Separate Domains of a Bifunctional Fusion Protein. Protein Eng. Des. Sel. 2001, 14, 529–532. [Google Scholar] [CrossRef]
- Robinson, C.R.; Sauer, R.T. Optimizing the Stability of Single-Chain Proteins by Linker Length and Composition Mutagenesis. Proc. Natl. Acad. Sci. USA 1998, 95, 5929–5934. [Google Scholar] [CrossRef]









| Group of Mice | M2e-Specific Tem Cells | Virus-Specific Tem Cells | ||
|---|---|---|---|---|
| CD4+ | CD8+ | CD4+ | CD8+ | |
| Experimental | 0.28 ± 0.07% | 0.276 ± 0.11% | 5.98 ± 0.63% | 13.69 ± 1.74% |
| Control (PBS) | 0.04 ± 0.02% | 0.08 ± 0.05% | 4.37 ± 0.63% | 8.60 ± 1.00% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zykova, A.A.; Blokhina, E.A.; Stepanova, L.A.; Shuklina, M.A.; Ozhereleva, O.O.; Tsybalova, L.M.; Kuprianov, V.V.; Ravin, N.V. Nanoparticles Carrying Conserved Regions of Influenza A Hemagglutinin, Nucleoprotein, and M2 Protein Elicit a Strong Humoral and T Cell Immune Response and Protect Animals from Infection. Molecules 2023, 28, 6441. https://doi.org/10.3390/molecules28186441
Zykova AA, Blokhina EA, Stepanova LA, Shuklina MA, Ozhereleva OO, Tsybalova LM, Kuprianov VV, Ravin NV. Nanoparticles Carrying Conserved Regions of Influenza A Hemagglutinin, Nucleoprotein, and M2 Protein Elicit a Strong Humoral and T Cell Immune Response and Protect Animals from Infection. Molecules. 2023; 28(18):6441. https://doi.org/10.3390/molecules28186441
Chicago/Turabian StyleZykova, Anna A., Elena A. Blokhina, Liudmila A. Stepanova, Marina A. Shuklina, Olga O. Ozhereleva, Liudmila M. Tsybalova, Victor V. Kuprianov, and Nikolai V. Ravin. 2023. "Nanoparticles Carrying Conserved Regions of Influenza A Hemagglutinin, Nucleoprotein, and M2 Protein Elicit a Strong Humoral and T Cell Immune Response and Protect Animals from Infection" Molecules 28, no. 18: 6441. https://doi.org/10.3390/molecules28186441
APA StyleZykova, A. A., Blokhina, E. A., Stepanova, L. A., Shuklina, M. A., Ozhereleva, O. O., Tsybalova, L. M., Kuprianov, V. V., & Ravin, N. V. (2023). Nanoparticles Carrying Conserved Regions of Influenza A Hemagglutinin, Nucleoprotein, and M2 Protein Elicit a Strong Humoral and T Cell Immune Response and Protect Animals from Infection. Molecules, 28(18), 6441. https://doi.org/10.3390/molecules28186441