Non-Targeted Spectranomics for the Early Detection of Xylella fastidiosa Infection in Asymptomatic Olive Trees, cv. Cellina di Nardò
Abstract
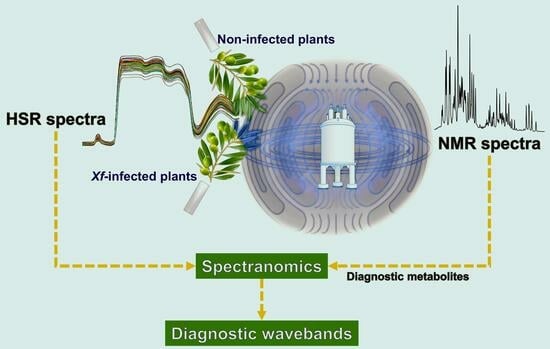
:1. Introduction
2. Results
2.1. Selection of Asymptomatic Leaves and qPCR Assay for Diagnosis of Xylella fastidiosa subsp. pauca ST53
2.2. Metabolic Profile from NMR Spectral Analysis
2.3. HSR Analysis
2.4. Chemometric Analysis of NMR Data
Correlation of NMR Diagnostics Signals to HSR
3. Discussion
4. Materials and Methods
4.1. Cultivation of Bacteria and Fungi
4.2. Cultivation and Artificial Infection of the Olive Plants
4.3. Diagnosis of Xylella fastidiosa subsp. pauca ST5 (Xf) in Inoculated Plants using qPCR Assay
4.4. Hyperspectral Reflectance (HSR)
4.5. NMR Sample Preparation and Spectra Acquisition
4.6. Chemometric Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saponari, M.; Boscia, D.; Nigro, F.; Martelli, G.P. Identification of DNA sequences related to Xylella fastidiosa in Oleander, almond and olive trees exhibiting leaf scorch symptoms in Apulia (Southern Italy). J. Plant Pathol. 2013, 95, 668. [Google Scholar] [CrossRef]
- Giampetruzzi, A.; D’Attoma, G.; Zicca, S.; Kubaa, R.A.; Rizzo, D.; Boscia, D.; Saldarelli, P.; Saponari, M. Draft genome sequence resources of three strains (tos4, tos5, and tos14) of Xylella fastidiosa infecting different host plants in the newly discovered outbreak in Tuscany, Italy. Phytopathology 2019, 109, 1516–1518. [Google Scholar] [CrossRef] [PubMed]
- Giampetruzzi, A.; Saponari, M.; Almeida, R.P.P.; Essakhi, S.; Boscia, D.; Loconsole, G.; Saldarelli, P. Complete Genome Sequence of the Olive-Infecting Strain Xylella fastidiosa subsp. pauca De Donno. Genome Announc. 2017, 5, 4–5. [Google Scholar] [CrossRef] [PubMed]
- Saponari, M.; Boscia, D.; Altamura, G.; Loconsole, G.; Zicca, S.; D’Attoma, G.; Morelli, M.; Palmisano, F.; Saponari, A.; Tavano, D.; et al. Isolation and pathogenicity of Xylella fastidiosa associated to the olive quick decline syndrome in southern Italy. Sci. Rep. 2017, 7, 17723. [Google Scholar] [CrossRef]
- Martelli, G.P.; Boscia, D.; Porcelli, F.; Saponari, M. The olive quick decline syndrome in south-east Italy: A threatening phytosanitary emergency. Eur. J. Plant Pathol. 2016, 144, 235–243. [Google Scholar] [CrossRef]
- Schneider, K.; van der Werf, W.; Cendoya, M.; Mourits, M.; Navas-Cortés, J.A.; Vicent, A.; Lansink, A.O. Impact of Xylella fastidiosa subspecies pauca in European olives. Proc. Natl. Acad. Sci. USA 2020, 117, 9250–9259. [Google Scholar] [CrossRef] [PubMed]
- Sabella, E.; Moretti, S.; Gärtner, H.; Luvisi, A.; De Bellis, L.; Vergine, M.; Saurer, M.; Cherubini, P. Increase in ring width, vessel number and δ18O in olive trees infected with Xylella fastidiosa. Tree Physiol. 2020, 40, 1583–1594. [Google Scholar] [CrossRef]
- Saponari, M.; Giampetruzzi, A.; Loconsole, G.; Boscia, D.; Saldarelli, P. Xylella fastidiosa in olive in apulia: Where we stand. Phytopathology 2019, 109, 175–186. [Google Scholar] [CrossRef] [PubMed]
- OEPP/EPPO. EPPO A1 and A2 Lists of Pests Recommended for Regulation as Quarantine Pests; European and Mediterranean Plant Protection Organization: Paris, France, 2018. [Google Scholar]
- European Commission Commission Implementing Regulation (EU) 2020/1201 of 14 August 2020 as regards measures to prevent the introduction into and the spread within the Union of Xylella fastidiosa (Wells et al.). Off. J. Eur. Union 2020, L 269, 2–39.
- Eghbalnia, H.R.; Romero, P.R.; Westler, W.M.; Baskaran, K.; Ulrich, E.L.; Markley, J.L. Increasing rigor in NMR-based metabolomics through validated and open source tools. Curr. Opin. Biotechnol. 2017, 43, 56–61. [Google Scholar] [CrossRef]
- Hall, R.D. Plant Metabolomics in a Nutshell: Potential and Future Challenges. Annu. Plant Rev. Online 2018, 43, 1–24. [Google Scholar] [CrossRef]
- Bohnenkamp, D.; Kuska, M.T.; Mahlein, A.K.; Behmann, J. Hyperspectral signal decomposition and symptom detection of wheat rust disease at the leaf scale using pure fungal spore spectra as reference. Plant Pathol. 2019, 68, 1188–1195. [Google Scholar] [CrossRef]
- Nicolì, F.; Negro, C.; Nutricati, E.; Vergine, M.; Aprile, A.; Sabella, E.; Damiano, G.; De Bellis, L.; Luvisi, A. Accumulation of azelaic acid in Xylella fastidiosa-infected olive trees: A mobile metabolite for health screening. Phytopathology 2019, 109, 318–325. [Google Scholar] [CrossRef]
- Mahlein, A.K.; Kuska, M.T.; Behmann, J.; Polder, G.; Walter, A. Hyperspectral sensors and imaging technologies in phytopathology: State of the art. Annu. Rev. Phytopathol. 2018, 56, 535–558. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Verdebout, J.; Schmuck, G.; Andreoli, G.; Hosgood, B. Investigation of leaf biochemistry by statistics. Remote Sens. Environ. 1995, 54, 189–197. [Google Scholar] [CrossRef]
- Curran, P.J. Remote sensing of foliar chemistry. Remote Sens. Environ. 1989, 30, 271–278. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Airborne spectranomics: Mapping canopy chemical and taxonomic diversity in tropical forests. Front. Ecol. Environ. 2009, 7, 269–276. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Spectranomics: Emerging science and conservation opportunities at the interface of biodiversity and remote sensing. Glob. Ecol. Conserv. 2016, 8, 212–219. [Google Scholar] [CrossRef]
- Ustin, S.L.; Roberts, D.A.; Gamon, J.A.; Asner, G.P.; Green, R.O. Using imaging spectroscopy to study ecosystem processes and properties. Bioscience 2004, 54, 523–534. [Google Scholar] [CrossRef]
- Fine, P.V.A.; Salazar, D.; Martin, R.E.; Metz, M.R.; Misiewicz, T.M.; Asner, G.P. Exploring the links between secondary metabolites and leaf spectral reflectance in a diverse genus of Amazonian trees. Ecosphere 2021, 12, e03362. [Google Scholar] [CrossRef]
- Weingarten, E.; Martin, R.E.; Hughes, R.F.; Vaughn, N.R.; Shafron, E.; Asner, G.P. Early detection of a tree pathogen using airborne remote sensing. Ecol. Appl. 2022, 32, e2519. [Google Scholar] [CrossRef] [PubMed]
- Gallo, V.; Ragone, R.; Musio, B.; Todisco, S.; Rizzuti, A.; Mastrorilli, P.; Pontrelli, S.; Intini, N.; Scapicchio, P.; Triggiani, M.; et al. A Contribution to the Harmonization of Non-targeted NMR Methods for Data-Driven Food Authenticity Assessment. Food Anal. Methods 2020, 13, 530–541. [Google Scholar] [CrossRef]
- Ragone, R.; Todisco, S.; Triggiani, M.; Pontrelli, S.; Latronico, M.; Mastrorilli, P.; Intini, N.; Ferroni, C.; Musio, B.; Gallo, V. Development of a food class-discrimination system by non-targeted NMR analyses using different magnetic field strengths. Food Chem. 2020, 332, 127339. [Google Scholar] [CrossRef]
- Musio, B.; Ragone, R.; Todisco, S.; Rizzuti, A.; Latronico, M.; Mastrorilli, P.; Pontrelli, S.; Intini, N.; Scapicchio, P.; Triggiani, M.; et al. A community-built calibration system: The case study of quantification of metabolites in grape juice by qNMR spectroscopy. Talanta 2020, 214, 120855. [Google Scholar] [CrossRef]
- Musio, B.; Todisco, S.; Antonicelli, M.; Garino, C.; Arlorio, M.; Mastrorilli, P.; Latronico, M.; Gallo, V. Non-Targeted NMR Method to Assess the Authenticity of Saffron and Trace the Agronomic Practices Applied for Its Production. Appl. Sci. 2022, 12, 2583. [Google Scholar] [CrossRef]
- Musio, B.; Ahmed, E.M.F.M.H.; Antonicelli, M.; Chiapperini, D.; Dursi, O.; Grieco, F.; Latronico, M.; Mastrorilli, P.; Ragone, R.; Settanni, R.; et al. A spectroscopic study to assess heavy metals absorption by a combined hemp/spirulina system from contaminated soil. Environ. Adv. 2022, 7, 100144. [Google Scholar] [CrossRef]
- Jlilat, A.; Ragone, R.; Gualano, S.; Santoro, F.; Gallo, V.; Varvaro, L.; Mastrorilli, P.; Saponari, M.; Nigro, F.; D’Onghia, A.M. A non-targeted metabolomics study on Xylella fastidiosa infected olive plants grown under controlled conditions. Sci. Rep. 2021, 11, 1070. [Google Scholar] [CrossRef]
- Rizzuti, A.; Aguilera-Saez, L.; Santoro, F.; Valentini, F.; Gualano, S.; D’Onghia, A.M.; Gallo, V.; Mastrorilli, P.; Latronico, M. Detection of Erwinia amylovora in pear leaves using a combined approach by hyperspectral reflectance and nuclear magnetic resonance spectroscopy. Phytopathol. Mediterr. 2018, 57, 241–252. [Google Scholar] [CrossRef]
- Rizzuti, A.; Caliandro, R.; Gallo, V.; Mastrorilli, P.; Chita, G.; Latronico, M. A combined approach for characterisation of fresh and brined vine leaves by X-ray powder diffraction, NMR spectroscopy and direct infusion high resolution mass spectrometry. Food Chem. 2013, 141, 1908–1915. [Google Scholar] [CrossRef]
- Nigro, F.; Boscia, D.; Antelmi, I.; Ippolito, A. Fungal Species Associated With a Severe Decline of Olive in Southern Italy. J. Plant Pathol. 2013, 95, 668. [Google Scholar] [CrossRef]
- Carlucci, A.; Raimondo, M.L.; Cibelli, F.; Phillips, A.J.L.; Lops, F. Phaeoacremonium aleophilum associated with a decline of olives in southern Italy. Phytopathol. Mediterr. 2013, 52, 517–527. [Google Scholar]
- Ortmayr, K.; Causon, T.J.; Hann, S.; Koellensperger, G. Increasing selectivity and coverage in LC-MS based metabolome analysis. TrAC-Trends Anal. Chem. 2016, 82, 358–366. [Google Scholar] [CrossRef]
- Fraga-Corral, M.; Carpena, M.; Garcia-Oliveira, P.; Pereira, A.G.; Prieto, M.A.; Simal-Gandara, J. Analytical Metabolomics and Applications in Health, Environmental and Food Science. Crit. Rev. Anal. Chem. 2020, 52, 712–734. [Google Scholar] [CrossRef] [PubMed]
- Girelli, C.R.; Del Coco, L.; Scortichini, M.; Petriccione, M.; Zampella, L.; Mastrobuoni, F.; Cesari, G.; Bertaccini, A.; D’Amico, G.; Contaldo, N.; et al. Xylella fastidiosa and olive quick decline syndrome (CoDiRO) in Salento (southern Italy): A chemometric 1H NMR-based preliminary study on Ogliarola salentina and Cellina di Nardò cultivars. Chem. Biol. Technol. Agric. 2017, 4, 25. [Google Scholar] [CrossRef]
- Mourad, E.F.; Sarhan, M.S.; Daanaa, H.S.A.; Abdou, M.; Morsi, A.T.; Abdelfadeel, M.R.; Elsawey, H.; Nemr, R.; El-Tahan, M.; Hamza, M.A.; et al. Plant materials are sustainable substrates supporting new technologies of plant-only-based culture media for in vitro culturing of the plant microbiota. Microbes Environ. 2018, 33, 40–49. [Google Scholar] [CrossRef]
- Sarhan, M.S.; Mourad, E.F.; Nemr, R.A.; Abdelfadeel, M.R.; Daanaa, H.S.A.; Youssef, H.H.; Goda, H.A.; Hamza, M.A.; Fayez, M.; Eichler-Löbermann, B.; et al. An inoculum-dependent culturing strategy (IDC) for the cultivation of environmental microbiomes and the isolation of novel endophytic Actinobacteria. J. Antibiot. 2020, 73, 66–71. [Google Scholar] [CrossRef]
- Castro-moretti, F.R.; Gentzel, I.N.; Mackey, D.; Alonso, A.P. Metabolomics as an emerging tool for the study of plant–pathogen interactions. Metabolites 2020, 10, 52. [Google Scholar] [CrossRef]
- Roper, C.; Castro, C.; Ingel, B. Xylella fastidiosa: Bacterial parasitism with hallmarks of commensalism. Curr. Opin. Plant Biol. 2019, 50, 140–147. [Google Scholar] [CrossRef]
- Riefolo, C.; Antelmi, I.; Castrignanò, A.; Ruggieri, S.; Galeone, C.; Belmonte, A.; Muolo, M.R.; Ranieri, N.A.; Labarile, R.; Gadaleta, G.; et al. Assessment of the hyperspectral data analysis as a tool to diagnose Xylella fastidiosa in the asymptomatic leaves of olive plants. Plants 2021, 10, 683. [Google Scholar] [CrossRef]
- Mechri, B.; Tekaya, M.; Hammami, M.; Chehab, H. Root verbascoside and oleuropein are potential indicators of drought resistance in olive trees (Olea europaea L.). Plant Physiol. Biochem. 2019, 141, 407–414. [Google Scholar] [CrossRef]
- Bulotta, S.; Oliverio, M.; Russo, D.; Procopio, A. Biological Activity of Oleuropein and its Derivatives. In Natural Products; Springer: Berlin/Heidelberg, Germany, 2013; pp. 3605–3638. ISBN 9783642221446. [Google Scholar]
- Özcan, M.M.; Matthäus, B. A review: Benefit and bioactive properties of olive (Olea europaea L.) leaves. Eur. Food Res. Technol. 2017, 243, 89–99. [Google Scholar] [CrossRef]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef]
- Dias, M.C.; Pinto, D.C.G.A.; Freitas, H.; Santos, C.; Silva, A.M.S. The antioxidant system in Olea europaea to enhanced UV-B radiation also depends on flavonoids and secoiridoids. Phytochemistry 2020, 170, 112199. [Google Scholar] [CrossRef] [PubMed]
- Mechri, B.; Tekaya, M.; Hammami, M.; Chehab, H. Effects of drought stress on phenolic accumulation in greenhouse-grown olive trees (Olea europaea). Biochem. Syst. Ecol. 2020, 92, 104112. [Google Scholar] [CrossRef]
- Zeiss, D.R.; Piater, L.A.; Dubery, I.A. Hydroxycinnamate Amides: Intriguing Conjugates of Plant Protective Metabolites. Trends Plant Sci. 2020, 26, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Guiso, M.; Marra, C. Highlights in oleuropein aglycone structure. Nat. Prod. Res. 2005, 19, 105–109. [Google Scholar] [CrossRef] [PubMed]
- De Pascali, M.; Vergine, M.; Negro, C.; Greco, D.; Vita, F.; Sabella, E.; De Bellis, L.; Luvisi, A. Xylella fastidiosa and Drought Stress in Olive Trees: A Complex Relationship Mediated by Soluble Sugars. Biology 2022, 11, 112. [Google Scholar] [CrossRef] [PubMed]
- Jeandet, P.; Formela-Luboińska, M.; Labudda, M.; Morkunas, I. The Role of Sugars in Plant Responses to Stress and Their Regulatory Function during Development. Int. J. Mol. Sci. 2022, 23, 5161. [Google Scholar] [CrossRef]
- Rosa, M.; Prado, C.; Podazza, G.; Interdonato, R.; González, J.A.; Hilal, M.; Prado, F.E. Soluble sugars-metabolism, sensing and abiotic stress a complex network in the life of plants. Plant Signal. Behav. 2009, 4, 388–393. [Google Scholar] [CrossRef]
- Nabavi, S.M.; Šamec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; et al. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020, 38, 107316. [Google Scholar] [CrossRef]
- Jeandet, P.; Vannozzi, A.; Sobarzo-Sánchez, E.; Uddin, M.S.; Bru, R.; Martínez-Márquez, A.; Clément, C.; Cordelier, S.; Manayi, A.; Nabavi, S.F.; et al. Phytostilbenes as agrochemicals: Biosynthesis, bioactivity, metabolic engineering and biotechnology. Nat. Prod. Rep. 2021, 38, 1282–1329. [Google Scholar] [CrossRef] [PubMed]
- Morkunas, I.; Marczak, Ł.; Stachowiak, J.; Stobiecki, M. Sucrose-induced lupine defense against Fusarium oxysporum: Sucrose-stimulated accumulation of isoflavonoids as a defense response of lupine to Fusarium oxysporum. Plant Physiol. Biochem. 2005, 43, 363–373. [Google Scholar] [CrossRef]
- Morkunas, I.; Narona, D.; Nowak, W.; Samardakiewicz, S.; Remlein-Starosta, D. Cross-talk interactions of sucrose and Fusarium oxysporum in the phenylpropanoid pathway and the accumulation and localization of flavonoids in embryo axes of yellow lupine. J. Plant Physiol. 2011, 168, 424–433. [Google Scholar] [CrossRef]
- Siddiqui, H.; Sami, F.; Hayat, S. Glucose: Sweet or bitter effects in plants-a review on current and future perspective. Carbohydr. Res. 2020, 487, 107884. [Google Scholar] [CrossRef]
- Lemos, E.G.D.M.; Alves, L.M.C.; Campanharo, J.C. Genomics-based design of defined growth media for the plant pathogen Xylella fastidiosa. FEMS Microbiol. Lett. 2003, 219, 39–45. [Google Scholar] [CrossRef]
- Wulff, N.A.; Mariano, A.G.; Gaurivaud, P.; De Almeida Souza, L.C.; Virgílio, A.C.D.; Monteiro, P.B. Influence of culture medium pH on growth, aggregation, and biofilm formation of Xylella fastidiosa. Curr. Microbiol. 2008, 57, 127–132. [Google Scholar] [CrossRef]
- Skodra, C.; Michailidis, M.; Dasenaki, M.; Ganopoulos, I.; Thomaidis, N.S.; Tanou, G.; Molassiotis, A. Unraveling salt-responsive tissue-specific metabolic pathways in olive tree. Physiol. Plant. 2021, 173, 1643–1656. [Google Scholar] [CrossRef]
- Freitas, D.D.S.; Carlos, E.F.; Gil, M.C.S.D.S.; Vieira, L.G.E.; Alcantara, G.B. NMR-Based Metabolomic Analysis of Huanglongbing-Asymptomatic and -Symptomatic Citrus Trees. J. Agric. Food Chem. 2015, 63, 7582–7588. [Google Scholar] [CrossRef]
- Huang, X.Y.; Wang, C.K.; Zhao, Y.W.; Sun, C.H.; Hu, D.G. Mechanisms and regulation of organic acid accumulation in plant vacuoles. Hortic. Res. 2021, 8, 227. [Google Scholar] [CrossRef]
- Girelli, C.R.; Angilè, F.; Del Coco, L.; Migoni, D.; Zampella, L.; Marcelletti, S.; Cristella, N.; Marangi, P.; Scortichini, M.; Fanizzi, F.P. 1H-NMR metabolite fingerprinting analysis reveals a disease biomarker and a field treatment response in xylella fastidiosa subsp. Pauca-infected olive trees. Plants 2019, 8, 115. [Google Scholar] [CrossRef]
- Sun, X.; Han, G.; Meng, Z.; Lin, L.; Sui, N. Roles of malic enzymes in plant development and stress responses. Plant Signal. Behav. 2019, 14, e1644596. [Google Scholar] [CrossRef] [PubMed]
- Martens, H. Quantitative Big Data: Where chemometrics can contribute. J. Chemom. 2015, 29, 563–581. [Google Scholar] [CrossRef]
- Martens, H. Interpretable machine learning with an eye for the physics: Hyperspectral Vis/NIR “video” of drying wood analyzed by hybrid subspace modeling. NIR News 2021, 32, 24–32. [Google Scholar] [CrossRef]
- Martens, H. Causality, machine learning and human insight. Anal. Chim. Acta 2023, 1277, 341585. [Google Scholar] [CrossRef] [PubMed]
- Sibanda, M.; Mutanga, O.; Chimonyo, V.G.P.; Clulow, A.D.; Shoko, C.; Mazvimavi, D.; Dube, T.; Mabhaudhi, T. Application of drone technologies in surface water resources monitoring and assessment: A systematic review of progress, challenges, and opportunities in the global south. Drones 2021, 5, 84. [Google Scholar] [CrossRef]
- Calderón, R.; Navas-Cortés, J.A.; Lucena, C.; Zarco-Tejada, P.J. High-resolution airborne hyperspectral and thermal imagery for early detection of Verticillium wilt of olive using fluorescence, temperature and narrow-band spectral indices. Remote Sens. Environ. 2013, 139, 231–245. [Google Scholar] [CrossRef]
- Poblete, T.; Camino, C.; Beck, P.S.A.; Hornero, A.; Kattenborn, T.; Saponari, M.; Boscia, D.; Navas-Cortes, J.A.; Zarco-Tejada, P.J. Detection of Xylella fastidiosa infection symptoms with airborne multispectral and thermal imagery: Assessing bandset reduction performance from hyperspectral analysis. ISPRS J. Photogramm. Remote Sens. 2020, 162, 27–40. [Google Scholar] [CrossRef]
- Chen, Y.C.; Thennadil, S.N. Insights into information contained in multiplicative scatter correction parameters and the potential for estimating particle size from these parameters. Anal. Chim. Acta 2012, 746, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Rinnan, Å.; van den Berg, F.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC-Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Lu, J.; Zhou, M.; Gao, Y.; Jiang, H. Using hyperspectral imaging to discriminate yellow leaf curl disease in tomato leaves. Precis. Agric. 2018, 19, 379–394. [Google Scholar] [CrossRef]
- Impollonia, G.; Croci, M.; Martani, E.; Ferrarini, A.; Kam, J.; Trindade, L.M.; Clifton-Brown, J.; Amaducci, S. Moisture content estimation and senescence phenotyping of novel Miscanthus hybrids combining UAV-based remote sensing and machine learning. GCB Bioenergy 2022, 14, 639–656. [Google Scholar] [CrossRef]
- Danson, F.M.; Steven, M.D.; Malthus, T.J.; Clark, J.A. High-spectral resolution data for determining leaf water content. Int. J. Remote Sens. 1992, 13, 461–470. [Google Scholar] [CrossRef]
- Scortichini, M. The multi-millennial olive agroecosystem of salento (Apulia, Italy) threatened by Xylella fastidiosa subsp. Pauca: A working possibility of restoration. Sustainability 2020, 12, 6700. [Google Scholar] [CrossRef]
- Sun, P.; Grignetti, A.; Liu, S.; Casacchia, R.; Salvatori, R.; Pietrini, F.; Loreto, F.; Centritto, M. Associated changes in physiological parameters and spectral reflectance indices in olive (Olea europaea L.) leaves in response to different levels of water stress. Int. J. Remote Sens. 2008, 29, 1725–1743. [Google Scholar] [CrossRef]
- Wells, J.M.; Raju, B.C.; Hung, H.; Weisburg, W.G.; Mandelco-paul, L.; Brenner, D.O.N.J. Limited, Fastidious Plant Bacteria Related to Xanthomonas spp. Int. J. Syst. Bacteriol. 1987, 37, 136–143. [Google Scholar] [CrossRef]
- Koch, A.L. Turbidity measurements of bacterial cultures in some available commercial instruments. Anal. Biochem. 1970, 38, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Saponari, M.; Boscia, D.; Altamura, G.; D’Attoma, G.; Cavalieri, V.; Zicca, S.; Morelli, M.; Tavano, D.; Loconsole, G.; Susca, L.; et al. Pilot project on Xylella fastidiosa to reduce risk assessment uncertainties. EFSA Support. Publ. 2016, 13, 1013E. [Google Scholar] [CrossRef]
- Hill, B.L.; Purcell, A.H. Acquisition and retention of Xylella fastidiosa by an efficient vector, Graphocephala atropunctata. Phytopathology 1995, 85, 209–212. [Google Scholar] [CrossRef]
- OEPP/EPPO. PM 7/24 (4) Xylella fastidiosa. Bull. OEPP/EPPO Bull. 2019, 49, 175–227. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef]
- Loconsole, G.; Potere, O.; Boscia, D.; Altamura, G.; Djelouah, K.; Elbeaino, T.; Frasheri, D.; Lorusso, D.; Palmisano, F.; Pollastro, P.; et al. Detection of Xylella fastidiosa in olive trees by molecular and serological methods. J. Plant Pathol. 2014, 96, 7–14. [Google Scholar] [CrossRef]
- Harper, S.J.; Ward, L.I.; Clover, G.R.G. Development of LAMP and Real-Time PCR Methods for the Rapid Detection of Xylella fastidiosa for Quarantine and Field Applications. Phytopathology 2010, 100, 1282–1288. [Google Scholar] [CrossRef] [PubMed]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Nevius, T.A.; Pardue, H.L. Development and Preliminary Evaluation of Modified Savitzky-Golay Smoothing Functions. Anal. Chem. 1984, 56, 2249–2251. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; De Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Szymańska, E.; Saccenti, E.; Smilde, A.K.; Westerhuis, J.A. Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 2012, 8, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.D.; D’Auria, J.C.; Silva Ferreira, A.C.; Gibon, Y.; Kruszka, D.; Mishra, P.; van de Zedde, R. High-throughput plant phenotyping: A role for metabolomics? Trends Plant Sci. 2022, 27, 549–563. [Google Scholar] [CrossRef]
- Cavill, R.; Jennen, D.; Kleinjans, J.; Briedé, J.J. Transcriptomic and metabolomic data integration. Brief. Bioinform. 2016, 17, 891–901. [Google Scholar] [CrossRef]
- Kumar Bharti, S.; Roy, R. Quantitative 1H NMR spectroscopy. TrAC-Trends Anal. Chem. 2012, 35, 5–26. [Google Scholar] [CrossRef]
- Mayerhöfer, T.G.; Pahlow, S.; Popp, J. The Bouguer-Beer-Lambert Law: Shining Light on the Obscure. ChemPhysChem 2020, 21, 2029–2046. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development Environment for R 2021. Available online: https://posit.co/ (accessed on 1 August 2022).
- R Core Team. R: A Language and Environment for Statistical Computing 2021. Available online: https://www.r-project.org/ (accessed on 1 August 2022).




| N° Leaves | Negative | Positive | HSR Samples (One Leaf) | NMR Samples (~5 Leaves) | |
|---|---|---|---|---|---|
| Non-inoculated | 134 | 134 | 0 | 134 | 27 |
| Xf-inoculated | 146 | 77 | 69 | 146 | 28 |
| Compound ID | Compound | δ (ppm) | Multiplicity | J (Hz) |
|---|---|---|---|---|
| Alcohols | ||||
| 1 | Ethanol | 1.18 | t | 6.5 |
| 3.65 | q | 6.5 | ||
| 2 | Methanol | 3.33 | s | |
| Organic acids | ||||
| 3 | Lactic acid | 1.34 | d | 6.9 |
| 4.15 | q | 6.9 | ||
| 4 | Citric acid | 2.70 | d | 15.0 |
| 2.80 | d | 15.5 | ||
| 5 | Formic acid | 8.43 | s | |
| 6 | Malic acid | 2.54 | dd | 15.8, 8.7 |
| 2.77 | dd | 15.8, 3.9 | ||
| 4.35 | dd | 8.7, 3.9 | ||
| 7 | Quinic acid | 1.87 | dd | 13.4, 10.8 |
| 1.96 | m | |||
| 2.07 | m | |||
| 3.56 | dd | 9.3; 3.3 | ||
| 4.04 | m | |||
| 4.14 | q | 3.5 | ||
| Carbohydrates | ||||
| 8 | Glucose | 3.24 | dd | 9.2, 7.9 |
| 3.43 | m | |||
| 3.48 | m | |||
| 3.53 | dd | 9.8, 3.8 | ||
| 3.75 | m | |||
| 3.83 | m | |||
| 3.87 | qd | 11.8, 2.4 | ||
| 4.65 | d | 7.9 | ||
| 5.23 | d | 3.7 | ||
| 9 | Mannitol | 3.68 | dd | 11.6; 6.2 |
| 3.77 | m | |||
| 3.81 | d | 8.6 | ||
| 3.88 | dd | 11.6; 2.5 | ||
| 10 | Fructose | 3.57 | m | |
| 3.71 | dd overlapped | |||
| 3.79 | m overlapped | |||
| 3.90 | dd overlapped | |||
| 4.00 | m | |||
| 4.03 | m | |||
| 4.11 | m | |||
| 11 | Sucrose | 3.48 | t | 9.2 |
| 3.57 | dd | 9.9; 3.7 | ||
| 3.67 | s | |||
| 3.78 | t | 9 | ||
| 3.83 | m | |||
| 3.87 | m | |||
| 3.91 | dd | 6.2; 3.5 | ||
| 4.05 | t | 8.5 | ||
| 4.22 | d | 8.7 | ||
| 5.42 | d | 3.8 | ||
| Amino Acids | ||||
| 12 | Alanine | 1.49 | d | 7.3 |
| 3.80 | q | 7.3 | ||
| 13 | Glycine | 3.53 | s | |
| Phenolic compounds | ||||
| 14 | Oleuropein derivatives | 1.85 | dd (methylenic proton of derivatives) | |
| 1.91 | ||||
| 6.67 | multiplets (aromatic protons of derivatives) | |||
| 6.79 | ||||
| 7.5 | ||||
| 8.95 | dd (aldehydic protons of the aglycone forms) | |||
| 9.02 | ||||
| 9.20 | ||||
| 9.21 | ||||
| 9.25 | ||||
| 15 | Tyrosol derivatives | 2.78 | t overlapped | |
| 3.78 | t overlapped | |||
| 6.94 | m | |||
| 6.75 | m | |||
| 7.12 | m | |||
| 7.14 | m | |||
| Quaternary ammonium compounds | ||||
| 16 | Choline | 3.20 | s | |
| 3.50 | dd overlapped | |||
| 4.05 | m | |||
| Fungi | Isolate Code | Non-Infected Plants (A) | Xf-Infected Plants (Xf) | ||
|---|---|---|---|---|---|
| Label | N Samples | Label | N Samples | ||
| Control | - | A-A | 5 | Xf-A | 7 |
| Phaeoacremonium aleophilum | B1a | A-F1 | 4 | Xf-F1 | 5 |
| Phaeoacremonium rubrigenum | N20 | A-F2 | 5 | Xf-F2 | 4 |
| Pseudophaeomoniella oleae | Fv84 | A-F3 | 4 | Xf-F3 | 4 |
| Pseudophaeomoniella oleicola | M24 | A-F4 | 3 | Xf-F4 | 5 |
| Pseudophaeomoniella oleicola | M51 | A-F5 | 6 | Xf-F5 | 3 |
| Total | 6 | 27 | 6 | 28 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, E.; Musio, B.; Todisco, S.; Mastrorilli, P.; Gallo, V.; Saponari, M.; Nigro, F.; Gualano, S.; Santoro, F. Non-Targeted Spectranomics for the Early Detection of Xylella fastidiosa Infection in Asymptomatic Olive Trees, cv. Cellina di Nardò. Molecules 2023, 28, 7512. https://doi.org/10.3390/molecules28227512
Ahmed E, Musio B, Todisco S, Mastrorilli P, Gallo V, Saponari M, Nigro F, Gualano S, Santoro F. Non-Targeted Spectranomics for the Early Detection of Xylella fastidiosa Infection in Asymptomatic Olive Trees, cv. Cellina di Nardò. Molecules. 2023; 28(22):7512. https://doi.org/10.3390/molecules28227512
Chicago/Turabian StyleAhmed, Elhussein, Biagia Musio, Stefano Todisco, Piero Mastrorilli, Vito Gallo, Maria Saponari, Franco Nigro, Stefania Gualano, and Franco Santoro. 2023. "Non-Targeted Spectranomics for the Early Detection of Xylella fastidiosa Infection in Asymptomatic Olive Trees, cv. Cellina di Nardò" Molecules 28, no. 22: 7512. https://doi.org/10.3390/molecules28227512
APA StyleAhmed, E., Musio, B., Todisco, S., Mastrorilli, P., Gallo, V., Saponari, M., Nigro, F., Gualano, S., & Santoro, F. (2023). Non-Targeted Spectranomics for the Early Detection of Xylella fastidiosa Infection in Asymptomatic Olive Trees, cv. Cellina di Nardò. Molecules, 28(22), 7512. https://doi.org/10.3390/molecules28227512