New Meroterpenoid Derivatives from the Pomegranate-Derived Endophytic Fungus Talaromyces purpureogenus
Abstract
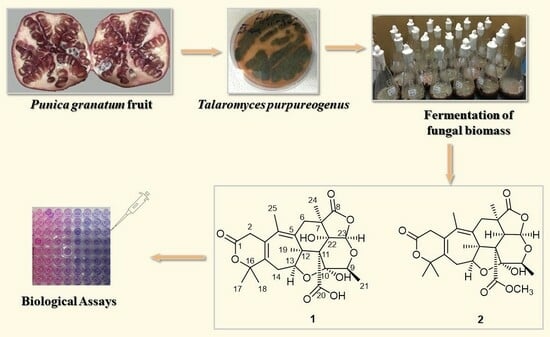
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material and Identification
3.3. Fermentation, Fractionation, and Isolation of Bioactive Compounds
3.4. Antimicrobial Susceptibility Tests
3.5. Computational Section
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef]
- Bogas, A.C.; Cruz, F.P.N.; Lacava, P.T.; Sousa, C.P. Endophytic fungi: An overview on biotechnological and agronomic potential. Braz. J. Biol. 2022, 84, e258557. [Google Scholar]
- Lee, C.; Shim, S.H. Endophytic Fungi Inhabiting Medicinal Plants and Their Bioactive Secondary Metabolites. Nat. Prod. Sci. 2020, 26, 10–27. [Google Scholar] [CrossRef]
- Gupta, S.; Chaturvedi, P.; Kulkarni, M.G.; Van Staden, J. A critical review on exploiting the pharmaceutical potential of plant endophytic fungi. Biotechnol. Adv. 2020, 39, 107462. [Google Scholar] [CrossRef] [PubMed]
- Digra, S.; Nonzom, S. An insight into endophytic antimicrobial compounds: An updated analysis. Plant Biotechnol. Rep. 2023, 17, 427–457. [Google Scholar]
- Samson, R.; Yilmaz, N.; Houbraken, J.; Spierenburg, H.; Seifert, K.; Peterson, S.; Varga, J.; Frisvad, J. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud. Mycol. 2011, 70, 159–183. [Google Scholar] [CrossRef]
- Feng, L.-X.; Zhang, B.-Y.; Zhu, H.-J.; Pan, L.; Cao, F. Bioactive Metabolites from Talaromyces purpureogenus, an Endophytic Fungus from Panax notoginseng. Chem. Nat. Compd. 2020, 56, 974–976. [Google Scholar] [CrossRef]
- Cao, X.; Shi, Y.; Wu, S.; Wu, X.; Wang, K.; Sun, H.; He, S.; Dickschat, J.S.; Wu, B. Polycyclic meroterpenoids, talaromyolides E − K for antiviral activity against pseudorabies virus from the endophytic fungus Talaromyces purpureogenus. Tetrahedron 2020, 76, 131349. [Google Scholar] [CrossRef]
- Do Nascimento, J.S.; Silva, F.M.; Magallanes-Noguera, C.A.; Kurina-Sanz, M.; dos Santos, E.G.; Caldas, I.S.; Luiz, J.H.H.; Silva, E.d.O. Natural trypanocidal product produced by endophytic fungi through co-culturing. Folia Microbiol. 2020, 65, 323–328. [Google Scholar] [CrossRef]
- Wang, W.; Wan, X.; Liu, J.; Wang, J.; Zhu, H.; Chen, C.; Zhang, Y. Two new terpenoids from Talaromyces purpurogenus. Mar. Drugs 2018, 16, 150. [Google Scholar] [CrossRef]
- Padmathilake, K.G.E.; Bandara, H.M.S.K.H.; Qader, M.M.; Kumar, N.S.; Jayasinghe, L.; Masubuti, H.; Fujimoto, Y. Talarofuranone, a New Talaroconvolutin Analog from the Endophytic Fungus Talaromyces purpurogenus from Pouteria campechiana Seeds. Nat. Prod. Commun. 2017, 12, 489–490. [Google Scholar] [CrossRef]
- Pandit, S.G.; Puttananjaih, M.H.; Harohally, N.V.; Dhale, M.A. Functional attributes of a new molecule-2-hydroxymethyl-benzoic acid 2′-hydroxy-tetradecyl ester isolated from Talaromyces purpureogenus CFRM02. Food Chem. 2018, 255, 89–96. [Google Scholar] [CrossRef]
- Zhao, J.-Y.; Wang, X.-J.; Liu, Z.; Meng, F.-X.; Sun, S.-F.; Ye, F.; Liu, Y.-B. Nonadride and Spirocyclic Anhydride Derivatives from the Plant Endophytic Fungus Talaromyces purpurogenus. J. Nat. Prod. 2019, 82, 2953–2962. [Google Scholar] [CrossRef] [PubMed]
- Ogbonna, C.; Hideki, A.; Ogbonna, J. Isolation and identification of Talaromyces purpurogenus and preliminary studies on its pigment production potentials in solid state cultures. Afr. J. Biotechnol. 2017, 16, 672–682. [Google Scholar]
- Sarlah, D.; Petrovčič, J.; Ungarean, C.N. Recent Chemical Methodology Advances in the Total Synthesis of Meroterpenoids. Acta Chim. Slov. 2021, 68, 247–267. [Google Scholar]
- Paluka, J.; Kanokmedhakul, K.; Soytong, M.; Soytong, K.; Kanokmedhakul, S. Meroditerpene pyrone, tryptoquivaline and brasiliamide derivatives from the fungus Neosartorya pseudofischeri. Fitoterapia 2019, 137, 104257. [Google Scholar] [CrossRef] [PubMed]
- Shang, Z.-C.; Yang, M.-H.; Liu, R.-H.; Wang, X.-B.; Kong, L.-Y. New Formyl Phloroglucinol Meroterpenoids from the Leaves of Eucalyptus robusta. Sci. Rep. 2016, 6, 39815. [Google Scholar] [CrossRef]
- Guo, J.; Ran, H.; Zeng, J.; Liu, D.; Xin, Z. Tafuketide, a phylogeny-guided discovery of a new polyketide from Talaromyces funiculosus Salicorn 58. Appl. Microbiol. Biotechnol. 2016, 100, 5323–5338. [Google Scholar] [CrossRef]
- Cen, S.; Jia, J.; Ge, Y.; Ma, Y.; Li, X.; Wei, J.; Bai, Y.; Wu, X.; Song, J.; Bi, H.; et al. A new antibacterial 3,5-dimethylorsellinic acid-based meroterpene from the marine fungus Aspergillus sp. CSYZ-1. Fitoterapia 2021, 152, 104908. [Google Scholar] [CrossRef]
- Stierle, D.B.; Stierle, A.A.; Patacini, B.; McIntyre, K.; Girtsman, T.; Bolstad, E. Berkeleyones and related meroterpenes from a deep water acid mine waste fungus that inhibit the production of interleukin 1-β from induced inflammasomes. J. Nat. Prod. 2011, 74, 2273–2277. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Huang, L.; Liu, Y.; Toshmatov, Z.; Zhang, C.; Shao, H. Two phytotoxins isolated from the pathogenic fungus of the invasive weed Xanthium italicum. Chem. Biodivers. 2020, 17, e2000043. [Google Scholar] [CrossRef]
- Zhao, M.; Ruan, Q.; Pan, W.; Tang, Y.; Zhao, Z.; Cui, H. New polyketides and diterpenoid derivatives from the fungus Penicillium sclerotiorum GZU-XW03-2 and their anti-inflammatory activity. Fitoterapia 2020, 143, 104561. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, X.; Lin, Y.; Yuan, J.; Lu, Y.; Zhu, X.; Li, J.; Li, M.; Lin, Y.; He, J.; et al. Meroterpenes and azaphilones from marine mangrove endophytic fungus Penicillium 303#. Fitoterapia 2014, 97, 241–246. [Google Scholar]
- Mándi, A.; Kurtán, T. Applications of OR/ECD/VCD to the structure elucidation of natural products. Nat. Prod. Rep. 2019, 36, 889–918. [Google Scholar] [CrossRef]
- Superchi, S.; Scafato, P.; Gorecki, M.; Pescitelli, G. Absolute configuration determination by quantum mechanical calculation of chiroptical spectra: Basics and applications to fungal metabolites. Curr. Med. Chem. 2018, 25, 287–320. [Google Scholar] [CrossRef]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef]
- Fernebro, J. Fighting bacterial infections—Future treatment options. Drug Resist. Updates 2011, 14, 125–139. [Google Scholar] [CrossRef]
- Barceloux, D.G. Medical Toxicology of Natural Substances: Foods, Fungi, Medicinal Herbs, Plants, and Venomous Animals; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Mabhula, A.; Singh, V. Drug-resistance in Mycobacterium tuberculosis: Where we stand. Medchemcomm 2019, 10, 1342–1360. [Google Scholar] [CrossRef] [PubMed]
- Gagneux, S. Ecology and evolution of Mycobacterium tuberculosis. Nat. Rev. Microbiol. 2018, 16, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Kjer, J.; Debbab, A.; Aly, A.H.; Proksch, P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat. Protoc. 2010, 5, 479–490. [Google Scholar] [CrossRef]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard, 9th ed.; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2012. [Google Scholar]
- Rehberg, N.; Akone, H.S.; Ioerger, T.R.; Erlenkamp, G.; Daletos, G.; Gohlke, H.; Proksch, P.; Kalscheuer, R. Chlorflavonin Targets Acetohydroxyacid Synthase Catalytic Subunit IlvB1 for Synergistic Killing of Mycobacterium tuberculosis. ACS Infect. Dis. 2018, 4, 123–134. [Google Scholar] [CrossRef] [PubMed]
- MacroModel. Schrödinger LLC. 2015. Available online: https://www.schrodinger.com/products/macromodel (accessed on 1 October 2023).
- Chai, J.-D.; Head-Gordon, M. Systematic optimization of long-range corrected hybrid density functionals. J. Chem. Phys. 2008, 128, 084106. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision E.01; Gaussian Inc.: Wallingford, CT, USA, 2013. [Google Scholar]
- Stephens, P.J.; Harada, N. ECD cotton effect approximated by the Gaussian curve and other methods. Chirality 2010, 22, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Varetto, U. MOLEKEL, 5.4; Swiss National Supercomputing Centre: Manno, Switzerland, 2009. [Google Scholar]







| Position | δC 1 | δH1, Multiplicity (J in Hz) | δC 2 | δH2, Multiplicity (J in Hz) |
|---|---|---|---|---|
| 1 | 168.71, C | 169.11, C | ||
| 2 | 31.51, CH2 | β 3.34, d (21.2) | 31.74, CH2 | β 3.27, d (21.1) |
| α 2.93, d (21.2) | α 2.83, d (21.1) | |||
| 3 | 128.97, C | 128.57, C | ||
| 4 | 131.65, C | 129.57, C | ||
| 5 | 133.8, C | 137.35, C | ||
| 6 | 32.51, CH2 | β 2.89, d (16.2) | 42.54, CH2 | β 2.94, d (16.3) |
| α 2.26, d (16.2) | α 2.26, d (16.3) | |||
| 7 | 46.53, C | 41.2, C | ||
| 8 | 173.36, C | 173.36, C | ||
| 9 | 70.33, CH | 4.07, q (6.3) | 67.23, CH | 4.62, q (6.5) |
| 10 | 99.10, C | 109.34, C | ||
| 11 | 72.38, C | 57.14, C | ||
| 12 | 52.02, C | 51.12, C | ||
| 13 | 92.91, CH | 4.23, dd (13, 3.75) | 94.47, CH | 4.16, dd (13, 4.5) |
| 14 | 31.43, CH2 | β 2.39, dd (13, 3.75) | 33.75, CH2 | β 2.66, t (13) |
| α 2.20, d (13) | α 2.47, dd (13, 4.5) | |||
| 15 | 130.88, C | 131.25, C | ||
| 16 | 85.05, C | 85.2, C | ||
| 17 | 27.69, CH3 | 1.49, s | 27.65, CH3 | 1.48, s |
| 18 | 27.79, CH3 | 1.62, s | 27.47, CH3 | 1.56, s |
| 19 | 27.91, CH3 | 1.45, s | 27.12, CH3 | 1.07, s |
| 20 | 164.80, C | 169.82, C | ||
| 21 | 13.31, CH3 | 1.42, d (6.4) | 13.26, CH3 | 1.24, d (5.9) |
| 22 | 80.68, C | 48.78, CH | 2.85, s | |
| 23 | 98.53, CH | 5.97, s | 90.90, CH | 5.43, s |
| 24 | 18.64, CH3 | 1.38, s | 21.53, CH3 | 1.39, s |
| 25 | 17.89, CH3 | 1.90, s | 17.2, CH3 | 1.71, s |
| 26 | 51.3, CH3 | 3.68, s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anwar, A.; Elnaggar, M.S.; Elissawy, A.M.; Ibrahim, N.; Mándi, A.; Kurtán, T.; Liu, Z.; El-Ahmady, S.H.; Kalscheuer, R. New Meroterpenoid Derivatives from the Pomegranate-Derived Endophytic Fungus Talaromyces purpureogenus. Molecules 2023, 28, 7650. https://doi.org/10.3390/molecules28227650
Anwar A, Elnaggar MS, Elissawy AM, Ibrahim N, Mándi A, Kurtán T, Liu Z, El-Ahmady SH, Kalscheuer R. New Meroterpenoid Derivatives from the Pomegranate-Derived Endophytic Fungus Talaromyces purpureogenus. Molecules. 2023; 28(22):7650. https://doi.org/10.3390/molecules28227650
Chicago/Turabian StyleAnwar, Alaa, Mohamed S. Elnaggar, Ahmed M. Elissawy, Nehal Ibrahim, Attila Mándi, Tibor Kurtán, Zhen Liu, Sherweit H. El-Ahmady, and Rainer Kalscheuer. 2023. "New Meroterpenoid Derivatives from the Pomegranate-Derived Endophytic Fungus Talaromyces purpureogenus" Molecules 28, no. 22: 7650. https://doi.org/10.3390/molecules28227650
APA StyleAnwar, A., Elnaggar, M. S., Elissawy, A. M., Ibrahim, N., Mándi, A., Kurtán, T., Liu, Z., El-Ahmady, S. H., & Kalscheuer, R. (2023). New Meroterpenoid Derivatives from the Pomegranate-Derived Endophytic Fungus Talaromyces purpureogenus. Molecules, 28(22), 7650. https://doi.org/10.3390/molecules28227650