Non-Invasive Detection of Biomolecular Abundance from Fermentative Microorganisms via Raman Spectra Combined with Target Extraction and Multimodel Fitting
Abstract
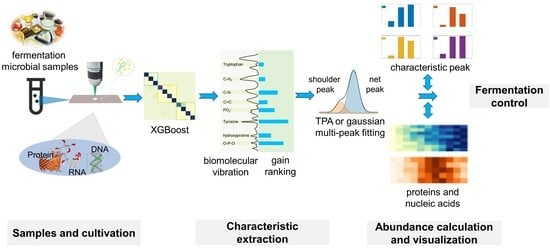
:1. Introduction
2. Results and Discussion
2.1. Characterization of the Bacterial Growth Process
2.2. Detection of the Growth Process of E. coli Using Raman Spectra
2.3. Target Extraction of Active Biomolecules during Cell Growth Process
2.4. Characteristic Peak Fitting to Improve the Accuracy of Biomolecular Abundance Calculations
2.5. Visual Analysis of Biological Abundance
3. Materials and Methods
3.1. Bacterium Culture
3.2. Raman Detection of Bacteria during the Growth Process
3.3. Data Pre-Processing
3.4. Target Extraction
3.5. Abundance Calculation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ren, Y.; Ji, Y.; Teng, L.; Zhang, H. Using Raman spectroscopy and chemometrics to identify the growth phase of Lactobacillus casei Zhang during batch culture at the single-cell level. Microb. Cell Factories 2017, 16, 233. [Google Scholar] [CrossRef]
- Luo, J.; Zhang, L.; Du, W.; Cheng, X.; Fang, F.; Cao, J.; Wu, Y.; Su, Y. Metagenomic approach reveals the fates and mechanisms of antibiotic resistance genes exposed to allicins during waste activated sludge fermentation: Insight of the microbial community, cellular status and gene regulation. Bioresour. Technol. 2021, 342, 125998. [Google Scholar] [CrossRef]
- Marx, V. A dream of single-cell proteomics. Nat. Methods 2019, 16, 809–812. [Google Scholar] [CrossRef]
- Zhang, L.; Vertes, A. Single-Cell Mass Spectrometry Approaches to Explore Cellular Heterogeneity. Angew. Chem. Int. Ed. 2018, 57, 4466–4477. [Google Scholar] [CrossRef]
- Jeon, J.; Cho, K.; Kang, J.; Park, S.; Ada, O.U.E.; Park, J.; Song, M.; Ly, Q.V.; Bae, H. Combined machine learning and biomolecular analysis for stability assessment of anaerobic ammonium oxidation under salt stress. Bioresour. Technol. 2022, 355, 127206. [Google Scholar] [CrossRef]
- Butler, H.J.; Ashton, L.; Bird, B.; Cinque, G.; Curtis, K.; Dorney, J.; Esmonde-White, K.; Fullwood, N.J.; Gardner, B.; Martin-Hirsch, P.L.; et al. Using Raman spectroscopy to characterize biological materials. Nat. Protoc. 2016, 11, 664–687. [Google Scholar] [CrossRef]
- He, Y.; Wang, X.; Ma, B.; Xu, J. Ramanome technology platform for label-free screening and sorting of microbial cell factories at single-cell resolution. Biotechnol. Adv. 2019, 37, 107388. [Google Scholar] [CrossRef]
- Jayan, H.; Pu, H.; Sun, D.W. Recent developments in Raman spectral analysis of microbial single cells: Techniques and applications. Crit. Rev. Food Sci. Nutr. 2022, 62, 4294–4308. [Google Scholar] [CrossRef]
- Masson, J.F.; Biggins, J.S.; Ringe, E. Machine learning for nanoplasmonics. Nat. Nanotechnol. 2023, 18, 111–123. [Google Scholar] [CrossRef]
- Siedhoff, N.E.; Schwaneberg, U.; Davari, M.D. Machine learning-assisted enzyme engineering. Methods Enzymol. 2020, 643, 281–315. [Google Scholar] [CrossRef]
- Shin, H. XGBoost Regression of the Most Significant Photoplethysmogram Features for Assessing Vascular Aging. IEEE J. Biomed. Health Inform. 2022, 26, 3354–3361. [Google Scholar] [CrossRef] [PubMed]
- Mojidra, R.; Hole, A.; Iwasaki, K.; Noothalapati, H.; Yamamoto, T.; Govekar, R. DNA Fingerprint Analysis of Raman Spectra Captures Global Genomic Alterations in Imatinib-Resistant Chronic Myeloid Leukemia: A Potential Single Assay for Screening Imatinib Resistance. Cells 2021, 10, 2506. [Google Scholar] [CrossRef] [PubMed]
- Wiemann, J.; Crawford, J.M.; Briggs, D.E.G. Phylogenetic and physiological signals in metazoan fossil biomolecules. Sci. Adv. 2020, 6, eaba6883. [Google Scholar] [CrossRef] [PubMed]
- Haga, Y.; Uemura, M.; Baba, S.; Inamura, K.; Takeuchi, K.; Nonomura, N.; Ueda, K. Identification of Multisialylated LacdiNAc Structures as Highly Prostate Cancer Specific Glycan Signatures on PSA. Anal. Chem. 2019, 91, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
- Talari, A.C.S.; Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2015, 50, 46–111. [Google Scholar] [CrossRef]
- Mueller, N.S.; Pfitzner, E.; Okamura, Y.; Gordeev, G.; Kusch, P.; Lange, H.; Heberle, J.; Schulz, F.; Reich, S. Surface-Enhanced Raman Scattering and Surface-Enhanced Infrared Absorption by Plasmon Polaritons in Three-Dimensional Nanoparticle Supercrystals. ACS Nano 2021, 15, 5523–5533. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Volponi, J.V.; Oliver, A.E.; Parikh, A.N.; Simmons, B.A.; Singh, S. In vivo lipidomics using single-cell Raman spectroscopy. Proc. Natl. Acad. Sci. USA 2011, 108, 3809–3814. [Google Scholar] [CrossRef]
- Ali, A.; Abouleila, Y.; Shimizu, Y.; Hiyama, E.; Watanabe, T.M.; Yanagida, T.; Germond, A. Single-Cell Screening of Tamoxifen Abundance and Effect Using Mass Spectrometry and Raman-Spectroscopy. Anal. Chem. 2019, 91, 2710–2718. [Google Scholar] [CrossRef]
- Willis, L.; Huang, K.C. Sizing up the bacterial cell cycle. Nat. Rev. Microbiol. 2017, 15, 606–620. [Google Scholar] [CrossRef]
- Reyes-Lamothe, R.; Sherratt, D.J. The bacterial cell cycle, chromosome inheritance and cell growth. Nat. Rev. Microbiol. 2019, 17, 467–478. [Google Scholar] [CrossRef]
- Shimizu, K.; Matsuoka, Y. Feedback regulation and coordination of the main metabolism for bacterial growth and metabolic engineering for amino acid fermentation. Biotechnol. Adv. 2022, 55, 107887. [Google Scholar] [CrossRef] [PubMed]
- Manjula-Basavanna, A.; Duraj-Thatte, A.M.; Joshi, N.S. Robust Self-Regeneratable Stiff Living Materials Fabricated from Microbial Cells. Adv. Funct. Mater. 2021, 31, 2010784. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hao, P.; Liu, K.; Shang, L.; Xu, L.; Lu, Z.; Li, B. Multi-point scanning confocal Raman spectroscopy for accurate identification of microorganisms at the single-cell level. Talanta 2022, 254, 124112. [Google Scholar] [CrossRef]
- Yang, K.; Li, H.Z.; Zhu, X.; Su, J.Q.; Ren, B.; Zhu, Y.G.; Cui, L. Rapid Antibiotic Susceptibility Testing of Pathogenic Bacteria Using Heavy-Water-Labeled Single-Cell Raman Spectroscopy in Clinical Samples. Anal. Chem. 2019, 91, 6296–6303. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.W.; Chen, Y.M.; Yao, B.Y.; Zhang, X.; Zeng, N.F. A neural network boosting regression model based on XGBoost. Appl. Soft Comput. 2022, 125, 109067. [Google Scholar] [CrossRef]
- Li, B.; Zhang, F.; Niu, Q.; Liu, J.; Yu, Y.; Wang, P.; Zhang, S.; Zhang, H.; Wang, Z. A molecular classification of gastric cancer associated with distinct clinical outcomes and validated by an XGBoost-based prediction model. Mol. Ther. Nucleic Acids 2022, 31, 224–240. [Google Scholar] [CrossRef]
- Ahmad, G.N.; Fatima, H.; Ullah, S.; Saidi, A.S. Efficient medical diagnosis of human heart diseases using machine learning techniques with and without GridSearchCV. IEEE Access 2022, 10, 80151–80173. [Google Scholar] [CrossRef]
- Xu, G.; Cheng, H.; Jones, R.; Feng, Y.; Gong, K.; Li, K.; Fang, X.; Tahir, M.A.; Valev, V.K.; Zhang, L. Surface-Enhanced Raman Spectroscopy Facilitates the Detection of Microplastics < 1 μm in the Environment. Environ. Sci. Technol. 2020, 54, 15594–15603. [Google Scholar] [CrossRef]
- Dastgeer, G.; Shahzad, Z.M.; Chae, H.; Kim, Y.H.; Ko, B.M.; Eom, J. Bipolar junction transistor exhibiting excellent output characteristics with a prompt response against the selective protein. Adv. Funct. Mater. 2022, 32, 2204781. [Google Scholar] [CrossRef]
- Mukherjee, R.; Verma, T.; Nandi, D.; Umapathy, S. Understanding the effects of culture conditions in bacterial growth: A biochemical perspective using Raman microscopy. J. Biophotonics 2020, 13, e201900233. [Google Scholar] [CrossRef]
- Kloß, S.; Rösch, P.; Pfister, W.; Kiehntopf, M.; Popp, J. Toward culture-free Raman spectroscopic identification of pathogens in ascitic fluid. Anal. Chem. 2015, 87, 937–943. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Li, S.; Wu, Q. Non-Invasive Detection of Biomolecular Abundance from Fermentative Microorganisms via Raman Spectra Combined with Target Extraction and Multimodel Fitting. Molecules 2024, 29, 157. https://doi.org/10.3390/molecules29010157
Li X, Li S, Wu Q. Non-Invasive Detection of Biomolecular Abundance from Fermentative Microorganisms via Raman Spectra Combined with Target Extraction and Multimodel Fitting. Molecules. 2024; 29(1):157. https://doi.org/10.3390/molecules29010157
Chicago/Turabian StyleLi, Xinli, Suyi Li, and Qingyi Wu. 2024. "Non-Invasive Detection of Biomolecular Abundance from Fermentative Microorganisms via Raman Spectra Combined with Target Extraction and Multimodel Fitting" Molecules 29, no. 1: 157. https://doi.org/10.3390/molecules29010157
APA StyleLi, X., Li, S., & Wu, Q. (2024). Non-Invasive Detection of Biomolecular Abundance from Fermentative Microorganisms via Raman Spectra Combined with Target Extraction and Multimodel Fitting. Molecules, 29(1), 157. https://doi.org/10.3390/molecules29010157