Ruthenium–Cyclopentadienyl–Cycloparaphenylene Complexes: Sizable Multicharged Cations Exhibiting High DNA-Binding Affinity and Remarkable Cytotoxicity
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis and Characterization of [(η6–-[12]CPP)[Ru(η5–-Cp)]12](PF6)12 and [(η6–-[11]CPP)[Ru(η5–-Cp)]11](PF6)11
2.2. Stability Studies of the Complex in Aqueous Media
2.3. Biological Studies
2.3.1. Fluorescence Quenching Studies of the d(5′-CGCGAATTCGCG-3′)2-Ethidium Bromide Adducts, with (2)
2.3.2. Cytotoxic Activity
3. Experimental
3.1. Materials and Methods
3.2. Preparation of Ligands and Complexes
3.3. Stability Studies of the Complex in Aqueous Media
3.4. Fluorescence Measurements
3.5. Cell Culture
3.6. Cell Growth Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Segawa, Y.; Fukazawa, A.; Matsuura, S.; Omachi, H.; Yamaguchi, S.; Irle, S.; Itami, K. Combined Experimental and Theoretical Studies on the Photophysical Properties of Cycloparaphenylenes. Org. Biomol. Chem. 2012, 10, 5979. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Zhuang, G.; Jia, H.; Wang, J.; Huang, Q.; Cui, S.; Du, P. A Novel Symmetrically Multifunctionalized Dodecamethoxy-Cycloparaphenylene: Synthesis, Photophysical, and Supramolecular Properties. Org. Chem. Front. 2018, 5, 1446–1451. [Google Scholar] [CrossRef]
- Darzi, E.R.; Sisto, T.J.; Jasti, R. Selective Syntheses of [7]-[12]Cycloparaphenylenes Using Orthogonal Suzuki-Miyaura Cross-Coupling Reactions. J. Org. Chem. 2012, 77, 6624–6628. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.K.; Kayahara, E.; Yamago, S. Practical Synthesis of [n]Cycloparaphenylenes (N=5, 7-12) by H2SnCl4-Mediated Aromatization of 1,4-Dihydroxycyclo-2,5-Diene Precursors. Chem.—Eur. J. 2015, 21, 5742–5749. [Google Scholar] [CrossRef] [PubMed]
- Kayahara, E.; Cheng, Y.; Yamago, S. Short-Step Synthesis of Large Cycloparaphenylenes. Chem. Lett. 2018, 47, 1108–1111. [Google Scholar] [CrossRef]
- Kayahara, E.; Sun, L.; Onishi, H.; Suzuki, K.; Fukushima, T.; Sawada, A.; Kaji, H.; Yamago, S. Gram-Scale Syntheses and Conductivities of [10]Cycloparaphenylene and Its Tetraalkoxy Derivatives. J. Am. Chem. Soc. 2017, 139, 18480–18483. [Google Scholar] [CrossRef] [PubMed]
- Lovell, T.C.; Colwell, C.E.; Zakharov, L.N.; Jasti, R. Symmetry Breaking and the Turn-on Fluorescence of Small, Highly Strained Carbon Nanohoops. Chem. Sci. 2019, 10, 3786–3790. [Google Scholar] [CrossRef] [PubMed]
- Darzi, E.R.; Hirst, E.S.; Weber, C.D.; Zakharov, L.N.; Lonergan, M.C.; Jasti, R. Synthesis, Properties, and Design Principles of Donor-Acceptor Nanohoops. ACS Cent. Sci. 2015, 1, 335–342. [Google Scholar] [CrossRef]
- Kuwabara, T.; Orii, J.; Segawa, Y.; Itami, K. Curved Oligophenylenes as Donors in Shape-Persistent Donor-Acceptor Macrocycles with Solvatofluorochromic Properties. Angew. Chem.—Int. Ed. 2015, 54, 9646–9649. [Google Scholar] [CrossRef]
- Tang, H.; Gu, Z.; Li, C.; Li, Z.; Wu, W.; Jiang, X. Nanoscale Vesicles Assembled from Non-Planar Cyclic Molecules for Efficient Cell Penetration. Biomater. Sci. 2019, 7, 2552–2558. [Google Scholar] [CrossRef]
- Lovell, T.C.; Bolton, S.G.; Kenison, J.P.; Shangguan, J.; Otteson, C.E.; Civitci, F.; Nan, X.; Pluth, M.D.; Jasti, R. Subcellular Targeted Nanohoop for One- and Two-Photon Live Cell Imaging. ACS Nano 2021, 15, 15285–15293. [Google Scholar] [CrossRef] [PubMed]
- White, B.M.; Zhao, Y.; Kawashima, T.E.; Branchaud, B.P.; Pluth, M.D.; Jasti, R. Expanding the Chemical Space of Biocompatible Fluorophores: Nanohoops in Cells. ACS Cent. Sci. 2018, 4, 1173–1178. [Google Scholar] [CrossRef]
- Adam, M.S.S.; Khalil, A. Bioreactivity of Divalent Bimetallic Vanadyl and Zinc Complexes Bis-Oxalyldihydrazone Ligand against Microbial and Human Cancer Series. CtDNA Interaction Mode. Int. J. Biol. Macromol. 2023, 249, 125917. [Google Scholar] [CrossRef] [PubMed]
- Adam, M.S.S. Zn(II)-Complexes of Diisatin Dihydrazones as Effective Catalysts in the Oxidation Protocol of Benzyl Alcohol and Effective Reagents for Biological Studies. J. Mol. Liq. 2023, 381, 121841. [Google Scholar] [CrossRef]
- Bresciani, G.; Busto, N.; Ceccherini, V.; Bortoluzzi, M.; Pampaloni, G.; Garcia, B.; Marchetti, F. Screening the Biological Properties of Transition Metal Carbamates Reveals Gold(I) and Silver(I) Complexes as Potent Cytotoxic and Antimicrobial Agents. J. Inorg. Biochem. 2022, 227, 111667. [Google Scholar] [CrossRef] [PubMed]
- Sifnaiou, E.; Tsolis, T.; Ypsilantis, K.; Roupakia, E.; Kolettas, E.; Plakatouras, J.C.; Garoufis, A. Synthesis and Characterization of Ruthenium-Paraphenylene-Cyclopentadienyl Full-Sandwich Complexes: Cytotoxic Activity against A549 Lung Cancer Cell Line and DNA Binding Properties. Molecules 2023, 29, 17. [Google Scholar] [CrossRef] [PubMed]
- Dale, L.D.; Tocher, J.H.; Dyson, T.M.; Edwards, D.I.; Tocher, D.A. Studies on DNA Damage and Induction of SOS Repair by Novel Multifunctional Bioreducible Compounds. II. A Metronidazole Adduct of a Ruthenium-Arene Compound. Anticancer Drug Des. 1992, 7, 3–14. [Google Scholar]
- Morris, R.E.; Aird, R.E.; del Socorro Murdoch, P.; Chen, H.; Cummings, J.; Hughes, N.D.; Parsons, S.; Parkin, A.; Boyd, G.; Jodrell, D.I.; et al. Inhibition of Cancer Cell Growth by Ruthenium(II) Arene Complexes. J. Med. Chem. 2001, 44, 3616–3621. [Google Scholar] [CrossRef]
- Tsolis, T.; Manos, M.J.; Karkabounas, S.; Zelovitis, I.; Garoufis, A. Synthesis, X-Ray Structure Determination, Cytotoxicity and Interactions with 9-Methylguanine, of Ruthenium(II) H6-Arene Complexes. J. Organomet. Chem. 2014, 768, 1–9. [Google Scholar] [CrossRef]
- Tsolis, T.; Nikolaou, N.; Ypsilantis, K.; Kougioumtzi, A.; Kordias, D.; Magklara, A.; Garoufis, A. Synthesis, Characterization, Interactions with 9-MeG and Cytotoxic Activity of Heterobimetallic RuII-PtII Complexes Bridged with 2, 2′-Bipyrimidine. J. Inorg. Biochem. 2021, 219, 111435. [Google Scholar] [CrossRef]
- Tsolis, T.; Ypsilantis, K.; Kourtellaris, A.; Garoufis, A. Synthesis, Characterization and Interactions with 9-Methylguanine of Ruthenium(II) H6-Arene Complexes with Aromatic Diimines. Polyhedron 2018, 149, 45–53. [Google Scholar] [CrossRef]
- Tsolis, T.; Papavasileiou, K.D.; Divanis, S.A.; Melissas, V.S.; Garoufis, A. How Half Sandwich Ruthenium Compounds Interact with DNA While Not Being Hydrolyzed; A Comparative Study. J. Inorg. Biochem. 2016, 160, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Loughrey, B.T.; Healy, P.C.; Parsons, P.G.; Williams, M.L. Selective Cytotoxic Ru(II) Arene Cp* Complex Salts [R-PhRuCp*]+X- for X = BF4-, PF6-, and BPh4-. Inorg. Chem. 2008, 47, 8589–8591. [Google Scholar] [CrossRef] [PubMed]
- Loughrey, B.T.; Williams, M.L.; Carruthers, T.J.; Parsons, P.G.; Healy, P.C. Synthesis, Structure, and Selective Cytotoxicity of Organometallic Cp *RuII O-Alkyl-N-Phenylcarbamate Sandwich Complexes. Aust. J. Chem. 2010, 63, 245–251. [Google Scholar] [CrossRef]
- Loughrey, B.T.; Cunning, B.V.; Healy, P.C.; Brown, C.L.; Parsons, P.G.; Williams, M.L. Selective, Cytotoxic Organoruthenium(II) Full-Sandwich Complexes: A Structural, Computational and in Vitro Biological Study. Chem.—Asian J. 2012, 7, 112–121. [Google Scholar] [CrossRef]
- Perekalin, D.S.; Molotkov, A.P.; Nelyubina, Y.V.; Anisimova, N.Y.; Kudinov, A.R. Synthesis of Amino Acid Esters of the Ruthenium Naphthalene Complex [(C5Me4CH2OH)Ru(C10H8)]+. Inorganica Chim. Acta 2014, 409, 390–393. [Google Scholar] [CrossRef]
- Perekalin, D.S.; Novikov, V.V.; Pavlov, A.A.; Ivanov, I.A.; Anisimova, N.Y.; Kopylov, A.N.; Volkov, D.S.; Seregina, I.F.; Bolshov, M.A.; Kudinov, A.R. Selective Ruthenium Labeling of the Tryptophan Residue in the Bee Venom Peptide Melittin. Chem.—Eur. J. 2015, 21, 4923–4925. [Google Scholar] [CrossRef]
- Bihari, Z.; Vultos, F.; Fernandes, C.; Gano, L.; Santos, I.; Correia, J.D.G.; Buglyó, P. Synthesis, Characterization and Biological Evaluation of a 67Ga-Labeled (H6-Tyr)Ru(H5-Cp) Peptide Complex with the HAV Motif. J. Inorg. Biochem. 2016, 160, 189–197. [Google Scholar] [CrossRef]
- Gelle, D.; Lamač, M.; Mach, K.; Šimková, L.; Gyepes, R.; Sommerová, L.; Martišová, A.; Bartošík, M.; Vaculovič, T.; Kanický, V.; et al. Enhanced Intracellular Accumulation and Cytotoxicity of Ferrocene-Ruthenium Arene Conjugates. Chempluschem 2020, 85, 1034–1043. [Google Scholar] [CrossRef]
- Matsui, K.; Segawa, Y.; Itami, K. Synthesis and Properties of Cycloparaphenylene-2,5-Pyridylidene: A Nitrogen-Containing Carbon Nanoring. Org. Lett. 2012, 14, 1888–1891. [Google Scholar] [CrossRef]
- Van Raden, J.M.; Louie, S.; Zakharov, L.N.; Jasti, R. 2,2′-Bipyridyl-Embedded Cycloparaphenylenes as a General Strategy To Investigate Nanohoop-Based Coordination Complexes. J. Am. Chem. Soc. 2017, 139, 2936–2939. [Google Scholar] [CrossRef] [PubMed]
- Heras Ojea, M.J.; Van Raden, J.M.; Louie, S.; Collins, R.; Pividori, D.; Cirera, J.; Meyer, K.; Jasti, R.; Layfield, R.A. Spin-Crossover Properties of an Iron(II) Coordination Nanohoop. Angew. Chem.—Int. Ed. 2021, 60, 3515–3518. [Google Scholar] [CrossRef] [PubMed]
- Kubota, N.; Segawa, Y.; Itami, K. H6-Cycloparaphenylene Transition Metal Complexes: Synthesis, Structure, Photophysical Properties, and Application to the Selective Monofunctionalization of Cycloparaphenylenes. J. Am. Chem. Soc. 2015, 137, 1356–1361. [Google Scholar] [CrossRef] [PubMed]
- Kayahara, E.; Patel, V.K.; Mercier, A.; Kündig, E.P.; Yamago, S. Regioselective Synthesis and Characterization of Multinuclear Convex-Bound Ruthenium-[ n ]Cycloparaphenylene (n =5 and 6) Complexes. Angew. Chem. Int. Ed. 2016, 55, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Ypsilantis, K.; Tsolis, T.; Garoufis, A. Interactions of (H5-CpRu)-[12]Cycloparaphenylene Full-Sandwich Complexes with 9-Methylguanine. Inorg. Chem. Commun. 2021, 134, 108992. [Google Scholar] [CrossRef]
- Bocekova-Gajdošíkova, E.; Epik, B.; Chou, J.; Akiyama, K.; Fukui, N.; Guénée, L.; Kündig, E.P. Microwave-Assisted Synthesis and Transformations of Cationic CpRu(II)(Naphthalene) and CpRu(II)(Naphthoquinone) Complexes. Helv. Chim. Acta 2019, 102, e1900076. [Google Scholar] [CrossRef]
- Evans, P.J.; Darzi, E.R.; Jasti, R. Efficient Room-Temperature Synthesis of a Highly Strained Carbon Nanohoop Fragment of Buckminsterfullerene. Nat. Chem. 2014, 6, 404–408. [Google Scholar] [CrossRef]
- Amaya, T.; Hirao, T. Chemistry of Sumanene. Chem. Rec. 2015, 15, 310–321. [Google Scholar] [CrossRef]
- Xia, A.; Selegue, J.P.; Carrillo, A.; Brock, C.P. Stereochemical Inversion of a Coordinated, Curved Hydrocarbon: Syntheses and Structures of exo- and endo-[Ru(η6-fluoradene)(η-C5Me5)][CF3SO3]. J. Am. Chem. Soc. 2000, 122, 3973–3974. [Google Scholar] [CrossRef]
- Kamieth, M.; Klärner, F.G.; Diederich, F. Modeling the Supramolecular Properties of Aliphatic-Aromatic Hydrocarbons with Convex-Concave Topology. Angew. Chem.—Int. Ed. 1998, 37, 3303–3306. [Google Scholar] [CrossRef]
- Ramana, M.M.V.; Betkar, R.; Nimkar, A.; Ranade, P.; Mundhe, B.; Pardeshi, S. In Vitro DNA Binding Studies of Antiretroviral Drug Nelfinavir Using Ethidium Bromide as Fluorescence Probe. J. Photochem. Photobiol. B Biol. 2015, 151, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Bittman, R. Studies of the Binding of Ethidium Bromide to Transfer Ribonucleic Acid: Absorption, Fluorescence, Ultracentrifugation and Kinetic Investigations. J. Mol. Biol. 1969, 46, 251–268. [Google Scholar] [CrossRef] [PubMed]
- Waring, M.J. Complex Formation between Ethidium Bromide and Nucleic Acids. J. Mol. Biol. 1965, 13, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Lepecq, J.-B.; Paoletti, C. A Fluorescent Complex between Ethidium Bromide and Nucleic Acids. J. Mol. Biol. 1967, 27, 87–106. [Google Scholar] [CrossRef]
- Galindo-Murillo, R.; Cheatham, T.E. Ethidium Bromide Interactions with DNA: An Exploration of a Classic DNA–Ligand Complex with Unbiased Molecular Dynamics Simulations. Nucleic Acids Res. 2021, 49, 3735–3747. [Google Scholar] [CrossRef] [PubMed]
- Rouzina, I.; Bloomfield, V.A. DNA Bending by Small, Mobile Multivalent Cations. Biophys. J. 1998, 74, 3152–3164. [Google Scholar] [CrossRef]
- Brukner, I.; Susic, S.; Dlakic, M.; Savic, A.; Pongor, S. Physiological Concentration of Magnesium Ions Induces a Strong Macroscopic Curvature in GGGCCC-Containing DNA. J. Mol. Biol. 1994, 236, 26–32. [Google Scholar] [CrossRef]
- Porschke, D. Structure and Dynamics of Double Helices in Solution: Modes of DNA Bending. J. Biomol. Struct. Dyn. 1986, 4, 373–389. [Google Scholar] [CrossRef]
- Goodsell, D.S.; Kopka, M.L.; Cascio, D.; Dickerson, R.E. Crystal Structure of CATGGCCATG and Its Implications for A-Tract Bending Models. Proc. Natl. Acad. Sci. USA 1993, 90, 2930–2934. [Google Scholar] [CrossRef]
- Marquet, R.; Houssier, C. Different Binding Modes of Spermine to A-T and G-C Base Pairs Modulate the Bending and Stiffening of the DNA Double Helix. J. Biomol. Struct. Dyn. 1988, 6, 235–246. [Google Scholar] [CrossRef]
- Duguid, J.; Bloomfield, V.A.; Benevides, J.; Thomas, G.J. Raman Spectroscopy of DNA-Metal Complexes. I. Interactions and Conformational Effects of the Divalent Cations: Mg, Ca, Sr, Ba, Mn, Co, Ni, Cu, Pd, and Cd. Biophys. J. 1993, 65, 1916–1928. [Google Scholar] [CrossRef] [PubMed]
- Rykowski, S.; Gurda-Woźna, D.; Orlicka-Płocka, M.; Fedoruk-Wyszomirska, A.; Giel-Pietraszuk, M.; Wyszko, E.; Kowalczyk, A.; Stączek, P.; Biniek-Antosiak, K.; Rypniewski, W.; et al. Design of DNA Intercalators Based on 4-Carboranyl-1,8-Naphthalimides: Investigation of Their DNA-Binding Ability and Anticancer Activity. Int. J. Mol. Sci. 2022, 23, 4598. [Google Scholar] [CrossRef] [PubMed]
- Kayahara, E.; Patel, V.K.; Xia, J.; Jasti, R.; Yamago, S. Selective and Gram-Scale Synthesis of [6]Cycloparaphenylene. Synlett 2015, 26, 1615–1619. [Google Scholar] [CrossRef]
- Yamago, S.; Watanabe, Y.; Iwamoto, T. Synthesis of [8]Cycloparaphenylene from a Square-Shaped Tetranuclear Platinum Complex. Angew. Chem. Int. Ed. 2010, 49, 757–759. [Google Scholar] [CrossRef]
- Rehman, S.U.; Yaseen, Z.; Husain, M.A.; Sarwar, T.; Ishqi, H.M.; Tabish, M. Interaction of 6 Mercaptopurine with Calf Thymus DNA—Deciphering the Binding Mode and Photoinduced DNA Damage. PLoS ONE 2014, 9, e93913. [Google Scholar] [CrossRef]







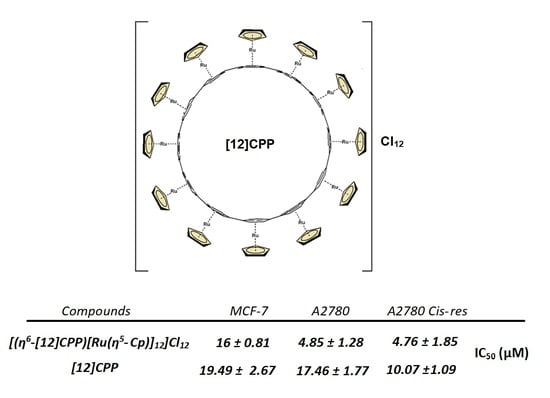
| Compounds | MCF-7 | A2780 | A2780 Cis-Res |
|---|---|---|---|
| (2) | 16 ± 0.81 | 4.85 ± 1.28 | 4.76 ± 1.85 |
| [12]CPP | 19.49 ± 2.67 | 17.46 ± 1.77 | 10.07 ± 1.09 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ypsilantis, K.; Sifnaiou, E.; Garypidou, A.; Kordias, D.; Magklara, A.; Garoufis, A. Ruthenium–Cyclopentadienyl–Cycloparaphenylene Complexes: Sizable Multicharged Cations Exhibiting High DNA-Binding Affinity and Remarkable Cytotoxicity. Molecules 2024, 29, 514. https://doi.org/10.3390/molecules29020514
Ypsilantis K, Sifnaiou E, Garypidou A, Kordias D, Magklara A, Garoufis A. Ruthenium–Cyclopentadienyl–Cycloparaphenylene Complexes: Sizable Multicharged Cations Exhibiting High DNA-Binding Affinity and Remarkable Cytotoxicity. Molecules. 2024; 29(2):514. https://doi.org/10.3390/molecules29020514
Chicago/Turabian StyleYpsilantis, Konstantinos, Evangelia Sifnaiou, Antonia Garypidou, Dimitrios Kordias, Angeliki Magklara, and Achilleas Garoufis. 2024. "Ruthenium–Cyclopentadienyl–Cycloparaphenylene Complexes: Sizable Multicharged Cations Exhibiting High DNA-Binding Affinity and Remarkable Cytotoxicity" Molecules 29, no. 2: 514. https://doi.org/10.3390/molecules29020514
APA StyleYpsilantis, K., Sifnaiou, E., Garypidou, A., Kordias, D., Magklara, A., & Garoufis, A. (2024). Ruthenium–Cyclopentadienyl–Cycloparaphenylene Complexes: Sizable Multicharged Cations Exhibiting High DNA-Binding Affinity and Remarkable Cytotoxicity. Molecules, 29(2), 514. https://doi.org/10.3390/molecules29020514