Comprehensive Investigation of Ginsenosides in the Steamed Panax quinquefolius with Different Processing Conditions Using LC-MS
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Ginsenosides in PQ Samples
2.2. Method Validation
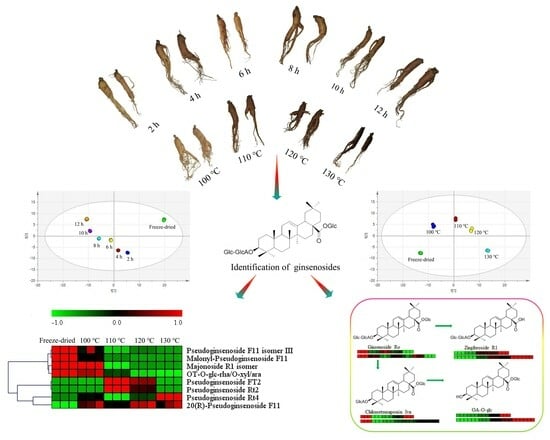
2.3. Difference between Steamed and Freeze-Dried Samples
2.4. Influence of Steam Temperature and Time on the Ginsenosides Composition
3. Materials and Methods
3.1. Chemical and Reagents
3.2. Sample Information and Preparation
3.3. LC-MS Analysis
3.4. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xiong, H.; Zhang, A.; Zhao, Q.; Yan, G.; Sun, H.; Wang, X. Discovery of quality-marker ingredients of Panax quinquefolius driven by high-throughput chinmedomics approach. Phytomedicine 2020, 74, 152928. [Google Scholar] [CrossRef]
- Yang, Z.; Deng, J.; Liu, M.; He, C.; Feng, X.; Liu, S.; Wei, S. A review for discovering bioactive minor saponins and biotransformative metabolites in Panax quinquefolius L. Front. Pharmacol. 2022, 13, 972813. [Google Scholar] [CrossRef]
- Liu, L.; Xu, F.; Wang, Y. Traditional uses, chemical diversity and biological activities of Panax L. (Araliaceae): A review. J. Ethnopharmacol. 2020, 263, 112792. [Google Scholar] [CrossRef]
- Hong, H.; Kim, J.; Lim, T.; Song, Y.; Cho, C.; Jang, M. Mixing ratio optimization for functional complex extracts of rhodiolacrenulata, Panax quinquefolius, and astragalus membranaceus using mixturedesign and verification of immune functional efficacy in animal models. J. Funct. Foods. 2018, 40, 447–454. [Google Scholar] [CrossRef]
- Wei, G.; Yang, F.; Wei, F.; Zhang, L.; Gao, Y.; Qian, J.; Chen, Z.; Jia, Z.; Wang, Y.; Su, H.; et al. Metabolomes and transcriptomes revealed the saponin distribution in root tissues of Panax quinquefolius and Panax notoginseng. J. Ginseng Res. 2020, 44, 757–769. [Google Scholar] [CrossRef]
- Huang, L.; Ren, C.; Li, H.; Wu, Y. Recent progress on processing technologies, chemical components, and bioactivities of Chinese red ginseng, American red ginseng, and Korean red ginseng. Food Bioprocess Technol. 2022, 15, 47–71. [Google Scholar] [CrossRef]
- Yoo, K.; Lee, C.; Lo, M.; Moon, B. The hypoglycemic effects of American red ginseng (Panax quinquefolius L.) on a diabetic mouse model. J. Food Sci. 2012, 77, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Bai, Y.; Huang, X.; Liu, X.; Cai, G.; Liu, S.; Guo, Y.; Gong, J. Comparison of the saponins in three processed American ginseng products by ultra-high performance liquid chromatography-quadrupole orbitrap tandem mass spectrometry and multivariate statistical analysis. Int. J. Anal. Chem. 2022, 2022, 6721937. [Google Scholar] [CrossRef]
- Wang, Y.; Li, H.; Li, Y.; Zhu, H.; Jin, Y. Identification of natural compounds targeting Annexin A2 with an anti-cancer effect. Protein Cell. 2018, 9, 568–579. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Xu, X.; Wang, X.; Wang, H.; Mi, Y.; Gao, X.; Guo, D.; Yang, W. Enhanced identification of ginsenosides simultaneously from seven Panax herbal extracts by data-dependent acquisition including a preferred precursor ions list derived from an in-house programmed virtual library. J. Agric. Food Chem. 2022, 70, 13796–13807. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, J.; Zuo, T.; Hu, Y.; Li, Z.; Wang, H.; Xu, X.; Yang, W.; Guo, D. Advances and challenges in ginseng research from 2011 to 2020: The phytochemistry, quality control, metabolism, and biosynthesis. Nat. Prod. Rep. 2022, 39, 875–909. [Google Scholar] [CrossRef]
- Chen, W.; Balan, P.; Popovich, D. Comparison of ginsenoside components of various tissues of new zealand forest-grown Asian ginseng (Panax Ginseng) and American ginseng (Panax Quinquefolium L.). Biomolecules 2020, 10, 372. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Fang, X.; Chen, D. Pharmacokinetics and bioavailability of ginsenoside Rb1 and Rg1 from Panax notoginseng in rats. J. Ethnopharmacol. 2003, 84, 187–192. [Google Scholar] [CrossRef]
- Fu, L.; Zhang, W.; Zhou, X.; Fu, J.; He, C. Tumor cell membrane-camouflaged responsive nanoparticles enable MRI-guided immuno-chemodynamic therapy of orthotopic osteosarcoma. Bioact. Mater. 2022, 17, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chiou, W.; Zhang, J. Comparison of the pharmacological effects of Panax ginseng and Panax quinquefolium. Acta Pharmacol. Sin. 2008, 29, 1103–1108. [Google Scholar] [CrossRef]
- Kim, S.; Kim, A. Anti-breast cancer activity of fine black ginseng (Panax ginseng Meyer) and ginsenoside Rg. J. Ginseng Res. 2015, 39, 125–134. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, D. The preparation of ginsenoside Rg5, its antitumor activity against breast cancer cells and its targeting of PI3K. Nutrients 2020, 12, 246. [Google Scholar] [CrossRef] [PubMed]
- Yue, G.; Lee, J.; Li, L.; Chan, K.; Wong, E.; Chan, J.; Fung, K.; Lui, V.; Chiu, P.; Lau, C. Andrographis paniculata elicits anti-invasion activities by suppressing TM4SF3 gene expression and by anoikis-sensitization in esophageal cancer cells. Am. J. Cancer Res. 2015, 5, 3570–3587. [Google Scholar] [PubMed]
- Deng, X.; Zhao, J.; Qu, L.; Duan, Z.; Fu, R.; Zhu, C.; Fan, D. Ginsenoside Rh4 suppresses aerobic glycolysis and the expression of PD-L1 via targeting AKT in esophageal cancer. Biochem. Pharmacol. 2020, 178, 114038. [Google Scholar] [CrossRef]
- Huang, L.; Li, H.; Wu, Y. Processing technologies, phytochemistry, bioactivities and applications of black ginseng-a novel manufactured ginseng product: A comprehensive review. Food Chem. 2023, 407, 134714. [Google Scholar] [CrossRef]
- Zheng, M.; Xu, F.; Li, Y.; Xi, X.; Cui, X.; Han, C.; Zhang, X. Study on transformation of ginsenosides in different methods. BioMed. Res. Int. 2017, 2017, 8601027. [Google Scholar] [CrossRef]
- Liu, Z.; Wen, X.; Wang, C.; Li, W.; Huang, W.; Xiao, J.; Ruan, C.; Yuan, C. Remarkable impact of amino acids on ginsenoside transformationfrom fresh ginseng to red ginseng. J. Ginseng Res. 2020, 44, 424–434. [Google Scholar] [CrossRef]
- Hu, J.; Xu, X.; Li, W.; Wang, Y.; Liu, Y.; Wang, Z.; Wang, Y. Ginsenoside Rk1 ameliorates paracetamol-induced hepatotoxicity inmice through inhibition of inflammation, oxidative stress, nitrativestress and apoptosis. J. Ginseng Res. 2019, 43, 10–19. [Google Scholar] [CrossRef]
- Wang, C.; Aung, H.; Ni, M.; Wu, J.; Tong, R.; Wicks, S.; He, T.; Yuan, C. Red American ginseng: Ginsenoside constituents and antiproliferative activities of heat-processed Panax quinquefolius roots. Planta Med. 2007, 73, 669–674. [Google Scholar] [CrossRef]
- Sun, B.; Xu, M.; Li, Z.; Wang, Y.; Sung, C. UPLC-Q-TOF-MS/MS analysis for steaming times-dependent profiling of steamed Panax quinquefolius and its ginsenosides transformations induced by repetitious steaming. J. Ginseng Res. 2012, 36, 277–290. [Google Scholar] [CrossRef]
- Huang, X.; Liu, Y.; Zhang, Y.; Li, S.; Yue, H.; Chen, C.; Liu, S. Multicomponent assessment and ginsenoside conversions of Panax quinquefolium L. roots before and after steaming by HPLC-MSn. J. Ginseng Res. 2019, 43, 27–37. [Google Scholar] [CrossRef]
- Fan, W.; Yang, Y.; Li, L.; Fan, L.; Wang, Z.; Yang, L. Mass spectrometry-based profiling and imaging strategy, a fit-for-purpose tool for unveiling the transformations of ginsenosides in Panax notoginseng during processing. Phytomedicine 2022, 103, 154223. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Sun, L.; Zhang, Z.; Guo, Y.; Liu, S. Profiling and multivariate statistical analysis of Panax ginseng based on ultra-high-performance liquid chromatography coupled with quadrupole-time-of-flight mass spectrometry. J. Pharm. Biomed. Anal. 2015, 107, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Guo, Y.; Wang, Y.; Li, M.; Li, K.; Liu, X.; Fang, C.; Luo, J. Metabolomic Analysis Reveals Nutritional Diversity among Three Staple Crops and Three Fruits. Foods 2022, 11, 550. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, D.; Sun, C.; Li, Y.; Lu, H.; Wang, X. Comprehensive lipidome and metabolome profiling investigations of Panax quinquefolius and application in different growing regions using liquid chromatography coupled with mass spectrometry. J. Agric. Food Chem. 2021, 69, 6710–6719. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Hu, Y.; Wu, W.; Ye, M.; Guo, D. Saponins in the genus Panax L. (Araliaceae): A systematic review of their chemical diversity. Phytochemistry 2014, 106, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Tung, N.; Song, G.; Nhiem, N.; Ding, Y.; Tai, B.; Jin, L.; Lim, C.; Hyun, J.; Park, C.; Kang, H.; et al. Dammarane-type saponins from the flower buds of Panax ginseng and their intracellular radical scavenging capacity. J. Agric. Food Chem. 2010, 58, 868–874. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Park, H.; Lee, D.; Jayakodi, M.; Kim, N.; Koo, H.; Lee, S.; Kim, Y.; Kwon, S.; Yang, T. Integrated transcriptomic and metabolomic analysis of five Panax ginseng cultivars reveals the dynamics of ginsenoside biosynthesis. Front. Plant Sci. 2017, 8, 1048. [Google Scholar] [CrossRef] [PubMed]
- Mao, Q.; Bai, M.; Xu, J.; Kong, M.; Zhu, L.; Zhu, H.; Wang, Q.; Li, S. Discrimination of leaves of Panax ginseng and P. quinquefolius by ultrahigh performance liquid chromatography quadrupole/time-of-flightmass spectrometry based metabolomics approach. J. Pharm. Biomed. Anal. 2014, 97, 129–140. [Google Scholar] [CrossRef]
- Xu, X.; Gao, Y.; Xu, S.; Liu, H.; Xue, X.; Zhang, Y.; Zhang, H.; Liu, M.; Xiong, H.; Lin, R.; et al. Remarkable impact of steam temperature on ginsenosides transformation from fresh ginseng to red ginseng. J. Ginseng Res. 2018, 42, 277–287. [Google Scholar] [CrossRef]
- Wu, W.; Qin, Q.; Guo, Y.; Sun, J.; Liu, S. Studies on the chemical transformation of 20(S)-protopanaxatriol (PPT)-type ginsenosides Re, Rg2, and Rf using rapid resolution liquid chromatography coupled with quadruple-time-of-flight mass spectrometry (RRLC-Q-TOF-MS). J. Agric. Food Chem. 2012, 60, 10007–10014. [Google Scholar] [CrossRef]






| No. | m/z | tR/min | Ion Adducts | Formula | Aglycone | Sugar Chains | Proposed Name | Type |
|---|---|---|---|---|---|---|---|---|
| 1 | 793.4388 | 29.4 | [M − H]− | C42H66O14 | 455 | O-176/O-162 | Chikusetsusaponin Iva | OA |
| 2 | 793.4388 | 33.8 | [M − H]− | C42H66O14 | 455 | O-162/O-176 | Chikusetsusaponin Iva isomer I | OA |
| 3 | 793.4388 | 24.1 | [M − H]− | C42H66O14 | 455 | O-176-162 | Zingibroside R1 | OA |
| 4 | 955.4896 | 22.3 | [M − H]− | C48H76O19 | 455 | O-162-162/O-176 | Ginsenoside Ro isomer | OA |
| 5 | 955.4896 | 21.4 | [M − H]− | C48H76O19 | 455 | O-176-162/O-162 | Ginsenoside Ro * | OA |
| 6 | 663.4108 | 39.8 | [M + HCOO]− | C36H58O8 | 455 | O-162 | OA-O-glc | OA |
| 7 | 699.4323 | 12.2 | [M + HCOO]− | C36H62O10 | 491 | O-162 | Pseudoginsenoside Rt4 isomer I | OT |
| 8 | 699.4323 | 15.2 | [M + HCOO]− | C36H62O10 | 491 | O-162 | Pseudoginsenoside Rt4 isomer II | OT |
| 9 | 699.4323 | 11.1 | [M + HCOO]− | C36H62O10 | 491 | O-162 | Pseudoginsenoside Rt4 | OT |
| 10 | 785.4693 | 14.0 | [M − H]− | C41H70O14 | 491 | O-162/O-132 | Presudoginsenoside FT2 | OT |
| 11 | 825.4634 | 26.1 | [M + HCOO]− | C42H68O13 | 455 | O-162-162 | OA-O-glc-glc | OT |
| 12 | 831.4743 | 13.8 | [M + HCOO]− | C41H70O14 | 491 | O-162-132 | Majonoside R2 | OT |
| 13 | 831.4743 | 10.4 | [M + HCOO]− | C41H70O14 | 491 | O-162-132 | Pseudoginsenoside Rt2 | OT |
| 14 | 845.4900 | 9.3 | [M + HCOO]− | C42H72O14 | 491 | O-162-146 | Pseudoginsenoside F11 isomer III | OT |
| 15 | 845.4900 | 11.0 | [M + HCOO]− | C42H72O14 | 491 | O-162-146 | Pseudoginsenoside F11 isomer I | OT |
| 16 | 845.4900 | 12.0 | [M + HCOO]− | C42H72O14 | 491 | O-162-146 | Pseudoginsenoside F11 isomer II | OT |
| 17 | 845.4900 | 15.0 | [M + HCOO]− | C42H72O14 | 491 | O-162-146 | 24(R)-Pseudoginsenoside F11 | OT |
| 18 | 845.4900 | 14.7 | [M + HCOO]− | C42H72O14 | 491 | O-162-146 | 24(S)-Pseudoginsenoside F11 * | OT |
| 19 | 861.4845 | 11.7 | [M + HCOO]− | C42H72O15 | 491 | O-162-162 | Majonoside R1 | OT |
| 20 | 861.4845 | 6.4 | [M + HCOO]− | C42H72O15 | 491 | O-162/O-162 | Majonoside R1 isomer | OT |
| 21 | 885.4851 | 17.8 | [M − H]− | C45H74O17 | 491 | O-162-146 | Malonyl-Pseudoginsenoside F11 | OT |
| 22 | 887.4991 | 21.2 | [M + HCOO]− | C44H74O15 | 491 | O-162-146 | 24(R)-Vinaginsenoside R1 | OT |
| 23 | 887.4991 | 20.6 | [M + HCOO]− | C44H74O15 | 491 | O-162-146 | 24(S)-Vinaginsenoside R1 | OT |
| 24 | 977.5322 | 7.2 | [M + HCOO]− | C47H80O18 | 491 | O-132/O-162-146 | OT-O-glc-rha-/O-xyl/ara | OT |
| 25 | 815.4793 | 19.8 | [M + HCOO]− | C41H70O13 | 491 | O-146/O-132 | OT-O-rha-xyl/ara | OT |
| 26 | 667.4426 | 41.6 | [M + HCOO]− | C36H62O8 | 459 | O-162 | 20(R)-Ginsenoside Rh2 * | PPD |
| 27 | 667.4426 | 40.9 | [M + HCOO]− | C36H62O8 | 459 | O-162 | 20(S)-Ginsenoside Rh2 * | PPD |
| 28 | 799.4845 | 37.8 | [M + HCOO]− | C41H70O12 | 459 | O-162-132 | 20(S)-Gypenoside XIII | PPD |
| 29 | 799.4845 | 38.6 | [M + HCOO]− | C41H70O12 | 459 | O-162-132 | 20(R)-Gypenoside XIII | PPD |
| 30 | 829.4943 | 34.1 | [M + HCOO]− | C42H72O13 | 459 | O-162-162 | 20(S)-Ginsenoside Rg3 * | PPD |
| 31 | 829.4943 | 34.6 | [M + HCOO]− | C42H72O13 | 459 | O-162-162 | 20(R)-Ginsenoside Rg3 * | PPD |
| 32 | 829.4943 | 37.5 | [M + HCOO]− | C42H72O13 | 459 | O-162-162 | Ginsenoside Rg3 isomer II | PPD |
| 33 | 829.4943 | 36.2 | [M + HCOO]− | C42H72O13 | 459 | O-162-162 | Ginsenoside Rg3 isomer I | PPD |
| 34 | 829.4943 | 27.6 | [M + HCOO]− | C42H72O13 | 459 | O-162-162 | Ginsenoside Rg3 isomer III | PPD |
| 35 | 869.489 | 35.8 | [M − H]− | C45H74O16 | 459 | O-162-162 | Malonyl-Ginsenoside Rg3 | PPD |
| 36 | 869.489 | 37.1 | [M − H]− | C45H74O16 | 459 | O-162-162 | Malonyl-Ginsenoside Rg3 isomer I | PPD |
| 37 | 869.489 | 37.5 | [M − H]− | C45H74O16 | 459 | O-162-162 | Malonyl-Ginsenoside Rg3 isomer II | PPD |
| 38 | 871.5054 | 38.0 | [M + HCOO]− | C44H74O14 | 459 | O-162-162 | Acetyl-Ginsenoside Rg3 | PPD |
| 39 | 961.5385 | 30.9 | [M + HCOO]− | C47H80O17 | 459 | O-162-132/O-162 | Gypenoside IX | PPD |
| 40 | 961.5385 | 29.2 | [M + HCOO]− | C47H80O17 | 459 | O-162-132/O-162 | Notoginsenoside Fe | PPD |
| 41 | 961.5385 | 30.3 | [M + HCOO]− | C47H80O17 | 459 | O-162-132/O-162 | Saponin Ia | PPD |
| 42 | 961.5385 | 27.2 | [M + HCOO]− | C47H80O17 | 459 | O-162-162/O-132 | Vinaginsenoside R17 | PPD |
| 43 | 991.5466 | 25.2 | [M + HCOO]− | C48H82O18 | 459 | O-162-162/O-162 | Gypenoside XVII | PPD |
| 44 | 991.5466 | 24.7 | [M + HCOO]− | C48H82O18 | 459 | O-162/O-162-162 | Ginsenoside Rd * | PPD |
| 45 | 1031.5432 | 26.0 | [M − H]− | C51H84O21 | 459 | O-162-162/O-162 | Malonyl-Ginsenoside Rd | PPD |
| 46 | 1031.5432 | 27.2 | [M − H]− | C51H84021 | 459 | O-162-162/O-162 | Malonyl-Ginsenoside Rd isomer I | PPD |
| 47 | 1031.5432 | 25.6 | [M − H]− | C51H84021 | 459 | O-162-162/O-162 | Malonyl-Ginsenoside Rd isomer II | PPD |
| 48 | 1031.5432 | 28.3 | [M − H]− | C51H84O21 | 459 | O-162-162/O-162 | Malonyl-Ginsenoside Rd isomer III | PPD |
| 49 | 1033.5566 | 30.8 | [M + HCOO]− | C50H84O19 | 459 | O-162-162/O-162 | Acetyl-Ginsenoside Rd isomer II | PPD |
| 50 | 1033.5566 | 34.0 | [M + HCOO]− | C50H84O19 | 459 | O-162-162/O-162 | Acetyl-Ginsenoside Rd isomer I | PPD |
| 51 | 1033.5566 | 29.3 | [M + HCOO]− | C50H84O19 | 459 | O-162-162/O-162 | Acetyl-Ginsenoside Rd | PPD |
| 52 | 1107.5945 | 22.6 | [M − H]− | C54H92O23 | 459 | O-162-162/O-162-162 | Ginsenoside Rb1 * | PPD |
| 53 | 1107.5945 | 22.9 | [M − H]− | C54H92O23 | 459 | O-162-162/O-162-162 | Ginsenoside Rb1 isomer I | PPD |
| 54 | 1123.5897 | 23.3 | [M + HCOO]− | C53H90O22 | 459 | O-162-132/O-162-162 | Ginsenoside Rb3 | PPD |
| 55 | 1123.5897 | 23.6 | [M + HCOO]− | C53H90O22 | 459 | O-162-132/O-162-162 | Ginsenoside Rc | PPD |
| 56 | 1123.5897 | 23.9 | [M + HCOO]− | C53H90O22 | 459 | O-162-132/O-162-162 | Ginsenoside Rb2 * | PPD |
| 57 | 1137.6053 | 24.0 | [M + HCOO]− | C54H92O22 | 459 | O-162-162/O-162-146 | PPD-O-glc-glc/O-glc-rha | PPD |
| 58 | 1163.5850 | 24.4 | [M − H]− | C56H92O25 | 459 | O-162-162/O-162-132 | Malonyl-Ginsenoside Rb2 | PPD |
| 59 | 1163.5850 | 23.7 | [M − H]− | C56H92O25 | 459 | O-162-162/O-162-132 | Malonyl-Ginsenoside Rb3 | PPD |
| 60 | 1163.5850 | 24.2 | [M − H]− | C56H92O25 | 459 | O-162-162/O-162-132 | Malonyl-Ginsenoside Rc | PPD |
| 61 | 1163.5850 | 24.7 | [M − H]− | C56H92O25 | 459 | O-162-162/O-162-132 | Malonyl-Ginsenoside Rb2 isomer | PPD |
| 62 | 1165.5992 | 27.0 | [M + HCOO]− | C55H92O23 | 459 | O-162-162/O-162-132 | Acetyl-Ginsenoside Rc | PPD |
| 63 | 1165.5992 | 25.9 | [M + HCOO]− | C55H92O23 | 459 | O-162-162/O-162-132 | Acetyl-Ginsenoside Rb3 | PPD |
| 64 | 1165.5992 | 30.0 | [M + HCOO]− | C55H92O23 | 459 | O-162-162/O-162-132 | Acetyl-Ginsenoside Rb2 | PPD |
| 65 | 1193.5959 | 24.6 | [M − H]− | C57H94O26 | 459 | O-162-162/O-162-162 | Malonyl-Ginsenoside Rb1 isomer II | PPD |
| 66 | 1193.5959 | 23.2 | [M − H]− | C57H94O26 | 459 | O-162-162/O-162-162 | Malonyl-Ginsenoside Rb1 | PPD |
| 67 | 1193.5959 | 24.1 | [M − H]− | C57H94O26 | 459 | O-162-162/O-162-162 | Malonyl-Ginsenoside Rb1 isomer I | PPD |
| 68 | 1195.6107 | 25.0 | [M + HCOO]− | C56H94O24 | 459 | O-162-162/O-162-162 | Acetyl-Ginsenoside Rb1 | PPD |
| 69 | 1195.6107 | 25.7 | [M + HCOO]− | C56H94O24 | 459 | O-162-162/O-162-162 | Acetyl-Ginsenoside Rb1 isomer I | PPD |
| 70 | 1195.6107 | 27.5 | [M + HCOO]− | C56H94O24 | 459 | O-162-162/O-162-162 | Acetyl-Ginsenoside Rb1 isomer II | PPD |
| 71 | 975.5534 | 30.4 | [M + HCOO]− | C48H82O17 | 459 | O-162-162/O-146 | PPD-O-glc-glc/O-rha | PPD |
| 72 | 975.5534 | 31.3 | [M + HCOO]− | C48H82O17 | 459 | O-162-162/O-146 | PPD-O-glc-glc/O-rha isomer I | PPD |
| 73 | 975.5534 | 29.9 | [M + HCOO]− | C48H82O17 | 459 | O-162-162/O-146 | PPD-O-glc-glc/O-rha isomer II | PPD |
| 74 | 825.4975 | 36.0 | [M + HCOO]− | C43H72O12 | 459 | O-162/O-162 | Acetyl-PPD-O-glc/O-glc | PPD |
| 75 | 631.3842 | 38.3 | [M − H]− | C36H56O9 | 475 | O-176 | PPT-O-glcA | PPT |
| 76 | 683.4366 | 15.5 | [M + HCOO]− | C36H62O9 | 475 | O-162 | Chikusetsusaponin L10 | PPT |
| 77 | 683.4366 | 17.0 | [M + HCOO]− | C36H62O9 | 475 | O-162 | Ginsenoside Rh1 isomer | PPT |
| 78 | 683.4366 | 19.3 | [M + HCOO]− | C36H62O9 | 475 | O-162 | 20(S)-Ginsenoside Rh1 * | PPT |
| 79 | 683.4366 | 20.7 | [M + HCOO]− | C36H62O9 | 475 | O-162 | 20(R)-Ginsenoside Rh1 | PPT |
| 80 | 683.4366 | 22.9 | [M + HCOO]− | C36H62O9 | 475 | O-162 | Ginsenoside F1 | PPT |
| 81 | 815.4793 | 20.4 | [M + HCOO]− | C41H70O13 | 475 | O-162-132 | Ginsenoside F5 | PPT |
| 82 | 815.4793 | 19.1 | [M + HCOO]− | C41H70O13 | 475 | O-162-132 | Ginsenoside F3 | PPT |
| 83 | 815.4793 | 17.4 | [M + HCOO]− | C41H70O13 | 475 | O-162-132 | Notoginsenoside R2 | PPT |
| 84 | 815.4793 | 11.5 | [M + HCOO]− | C41H70O13 | 475 | O-162/O-132 | PPT-O-xyl/ara/O-glc | PPT |
| 85 | 815.4793 | 18.1 | [M + HCOO]− | C41H70O13 | 475 | O-162/O-132 | PPT-O-xyl/ara/O-glc isomer | PPT |
| 86 | 829.4943 | 19.3 | [M + HCOO]− | C42H72O13 | 475 | O-162-146 | 20(S)-Ginsenoside Rg2 * | PPT |
| 87 | 829.4943 | 20.0 | [M + HCOO]− | C42H72O13 | 475 | O-162-146 | 20(R)-Ginsenoside Rg2 * | PPT |
| 88 | 829.4943 | 15.5 | [M + HCOO]− | C42H72O13 | 475 | O-162-146 | Ginsenoside Rg2 isomer I | PPT |
| 89 | 829.4943 | 16.3 | [M + HCOO]− | C42H72O13 | 475 | O-162-146 | Ginsenoside Rg2 isomer II | PPT |
| 90 | 845.4913 | 8.8 | [M + HCOO]− | C42H72O14 | 475 | O-162/O-162 | Ginsenoside Rg1 * | PPT |
| 91 | 845.4913 | 16.6 | [M + HCOO]− | C42H72O14 | 475 | O-162-162 | Ginsenoside Rf | PPT |
| 92 | 845.4913 | 21.9 | [M + HCOO]− | C42H72O14 | 475 | O-162/O-162 | Ginsenoside La | PPT |
| 93 | 869.4890 | 21.3 | [M − H]− | C45H74O16 | 475 | O-162-146 | Malonyl-Ginsenoside Rg2 | PPT |
| 94 | 885.4851 | 10.5 | [M − H]− | C45H74O17 | 475 | O-162-162 | Malonyl-Ginsenoside Rf | PPT |
| 95 | 887.4991 | 13.7 | [M + HCOO]− | C44H74O15 | 475 | O-162-162 | Acetyl-20(R)-Ginsenoside Rg2 | PPT |
| 96 | 887.4991 | 13 | [M + HCOO]− | C44H74O15 | 475 | O-162/O-162 | Acetyl-20(S)-Ginsenoside Rg1 | PPT |
| 97 | 887.4991 | 14.4 | [M + HCOO]− | C44H74O15 | 475 | O-162/O-162 | Acetyl-20(R)-Ginsenoside Rg1 | PPT |
| 98 | 961.5385 | 10.4 | [M + HCOO]− | C47H80O17 | 475 | O-162-146/O-132 | Cyclofoetoside A isomer | PPT |
| 99 | 961.5385 | 11.7 | [M + HCOO]− | C47H80O17 | 475 | O-162-146/O-132 | Cyclofoetoside A | PPT |
| 100 | 977.5322 | 17.8 | [M + HCOO]− | C47H80O18 | 475 | O-162/O-162-132 | Quinquenoside L3 | PPT |
| 101 | 977.5322 | 7.7 | [M + HCOO]− | C47H80O18 | 475 | O-162-132/O-162 | Ginsenoside Re4 | PPT |
| 102 | 977.5322 | 8.1 | [M + HCOO]− | C47H80O18 | 475 | O-162-132/O-162 | 20(S)-Quinquenoside L17 | PPT |
| 103 | 977.5322 | 8.4 | [M + HCOO]− | C47H80O18 | 475 | O-162-132/O-162 | 20(R)-Quinquenoside L17 | PPT |
| 104 | 991.5466 | 8.7 | [M + HCOO]− | C48H82O18 | 475 | O-162-146/O-162 | Ginsenoside Re isomer | PPT |
| 105 | 991.5466 | 27.5 | [M + HCOO]− | C48H82O18 | 475 | O-162/O-162-162 | Chikusetsusaponin FK1 | PPT |
| 106 | 991.5466 | 9.5 | [M + HCOO]− | C48H82O18 | 475 | O-162-146/O-162 | Ginsenoside Re * | PPT |
| 107 | 1007.5416 | 6.9 | [M + HCOO]− | C48H82O19 | 475 | O-162/O-162-162 | Ginsenoside Re1 | PPT |
| 108 | 1007.5416 | 13.4 | [M + HCOO]− | C48H82O19 | 475 | O-162/O-162-162 | Ginsenoside Re2 | PPT |
| 109 | 1007.5416 | 7.4 | [M + HCOO]− | C48H82O19 | 475 | O-162/O-162-162 | Ginsenoside Re3 | PPT |
| 110 | 1031.5432 | 10.7 | [M − H]− | C51H84021 | 475 | O-162-146/O-162 | Malonyl-Ginsenoside Re | PPT |
| 111 | 1031.5432 | 11.4 | [M − H]− | C51H84021 | 475 | O-162-146/O-162 | Malonyl-Ginsenoside Re isomer I | PPT |
| 112 | 1031.5432 | 12.0 | [M − H]− | C51H84021 | 475 | O-162-146/O-162 | Malonyl-Ginsenoside Re isomer II | PPT |
| 113 | 1031.5432 | 12.5 | [M − H]− | C51H84021 | 475 | O-162-146/O-162 | Malonyl-Ginsenoside Re isomer III | PPT |
| 114 | 1033.5566 | 15.4 | [M + HCOO]− | C50H84O19 | 475 | O-162-146/O-162 | Acetyl-Ginsenoside Re | PPT |
| 115 | 1033.5566 | 14.0 | [M + HCOO]− | C50H84O19 | 475 | O-162-146/O-162 | Acetyl-Ginsenoside Re isomer | PPT |
| 116 | 1139.5848 | 11.1 | [M + HCOO]− | C53H90O23 | 475 | O-162-162/O-162-132 | Floralginsenoside P | PPT |
| 117 | 1123.5897 | 7.9 | [M + HCOO]− | C53H90O22 | 475 | O-162-132/O-162-146 | Floralquinquenoside E | PPT |
| 118 | 799.4845 | 23.4 | [M + HCOO]− | C41H70O12 | 475 | O-146-132 | PPT-O-rha-xyl/ara | PPT |
| 119 | 975.5534 | 15.3 | [M + HCOO]− | C48H82O17 | 475 | O-162-146/O-146 | PPT-O-glc-rha/O-rha ** | PPT |
| 120 | 665.4265 | 28.3 | [M + HCOO]− | C36H60O8 | 457 | O-162 | Ginsenoside Rh4 | OTHER |
| 121 | 665.4265 | 27.0 | [M + HCOO]− | C36H60O8 | 457 | O-162 | Ginsenoside Pk3 | OTHER |
| 122 | 665.4265 | 26.7 | [M + HCOO]− | C36H60O8 | 457 | O-162 | Ginsenoside Pk3 isomer | OTHER |
| 123 | 665.4265 | 40.3 | [M + HCOO]− | C36H60O8 | 457 | O-162 | Ginsenoside Rh16 | OTHER |
| 124 | 665.4265 | 42.0 | [M + HCOO]− | C36H60O8 | 457 | O-162 | Ginsenoside Rh16 isomer | OTHER |
| 125 | 701.4475 | 8.6 | [M − H]− | C36H64O10 | 493 | O-162 | 20(S)-Ginsenoside Rh12 | OTHER |
| 126 | 701.4475 | 9.3 | [M − H]− | C36H64O10 | 493 | O-162 | 20(R)-Ginsenoside Rh12 | OTHER |
| 127 | 703.4207 | 17.2 | [M + HCOO]− | C35H62O11 | 495 | O-162 | Floralginsenoside Tb | OTHER |
| 128 | 717.4421 | 6.6 | [M + HCOO]− | C36H64O11 | 509 | O-162 | Dammarane-3,6,12,24,25-pentol, 20-(β-d-glucopyranosyloxy)-, (3β,6β,12β)-(ACI) isomer | OTHER |
| 129 | 717.4421 | 7.8 | [M + HCOO]− | C36H64O11 | 509 | O-162 | Dammarane-3,6,12,24,25-pentol, 20-(β-d-glucopyranosyloxy)-, (3β,6β,12β)-(ACI) | OTHER |
| 130 | 781.4734 | 40.7 | [M + HCOO]− | C41H68O11 | 441 | O-162-132 | Notoginsenoside ST11 | OTHER |
| 131 | 811.4839 | 25.4 | [M + HCOO]− | C42H70O12 | 457 | O-162-146 | Ginsenoside Rg6 isomer I | OTHER |
| 132 | 811.4839 | 25.8 | [M + HCOO]− | C42H70O12 | 457 | O-162-146 | Ginsenoside Rg6 isomer II | OTHER |
| 133 | 811.4839 | 26.4 | [M + HCOO]− | C42H70O12 | 457 | O-162-146 | Ginsenoside Rg6 | OTHER |
| 134 | 811.4839 | 38.6 | [M + HCOO]− | C42H70O12 | 441 | O-162-162 | Ginsenoside Rg5 isomer I | OTHER |
| 135 | 811.4839 | 39.5 | [M + HCOO]− | C42H70O12 | 441 | O-162-162 | Ginsenoside Rg5 * | OTHER |
| 136 | 811.4839 | 39.0 | [M + HCOO]− | C42H70O12 | 441 | O-162-162 | Ginsenoside Rg5 isomer II | OTHER |
| 137 | 827.4783 | 38.4 | [M + HCOO]− | C42H70O13 | 457 | O-162-162 | 20(R)-5,6-Didehydroginsenoside Rg3 | OTHER |
| 138 | 827.4783 | 32.2 | [M + HCOO]− | C42H70O13 | 457 | O-162-162 | 20(S)-5,6-Didehydroginsenoside Rg3 | OTHER |
| 139 | 827.4783 | 28.0 | [M + HCOO]− | C42H70O13 | 457 | O-162-162 | Ginsenoside Rh15 | OTHER |
| 140 | 847.5047 | 8.6 | [M + HCOO]− | C42H74O14 | 493 | O-162-146 | 20(S)-25-OH-Ginsenoside Rg2 | OTHER |
| 141 | 847.5047 | 23.3 | [M + HCOO]− | C42H74O14 | 477 | O-162-162 | 20(S)-25-OH-Ginsenoside Rg3 | OTHER |
| 142 | 847.5047 | 24.0 | [M + HCOO]− | C42H74O14 | 477 | O-162-162 | 20(R)-25-OH-Ginsenoside Rg3 | OTHER |
| 143 | 849.4843 | 6.5 | [M + HCOO]− | C41H72O15 | 509 | O-162/O-132 | β-d-Glucopyranoside, (3β,12β)-3,12,24,25-tetrahydroxy-20-(d-xylopyranosyloxy)dammaran-6-yl (ACI) | OTHER |
| 144 | 853.4949 | 42.0 | [M + HCOO]− | C44H72O13 | 441 | O-162-162 | Ginsenoside Rs4 | OTHER |
| 145 | 863.501 | 6.7 | [M + HCOO]− | C42H74O15 | 509 | O-162-146 | Quinquenoside L9 | OTHER |
| 146 | 975.5534 | 24.3 | [M + HCOO]− | C48H82O17 | 443 | O-162-162/O-162 | Vinaginsenoside R3 | OTHER |
| 147 | 989.5324 | 24.0 | [M + HCOO]− | C48H80O18 | 457 | O-162-162/O-162 | 5,6-Didehydroginsenoside Rd | OTHER |
| 148 | 989.5324 | 13.0 | [M + HCOO]− | C48H80O18 | 473 | O-162/O-162-146 | Ginsenoside Rh18 | OTHER |
| 149 | 989.5324 | 20.8 | [M + HCOO]− | C48H80O18 | 457 | O-162-162/O-162 | Quinquenoside L1 | OTHER |
| 150 | 1005.527 | 10.7 | [M + HCOO]− | C48H80O19 | 473 | O-162-162/O-162 | Vinaginsenoside R20 | OTHER |
| 151 | 1007.5416 | 6.3 | [M + HCOO]− | C48H82O19 | 491 | O-162-146/O-146 | Majoroside F5 | OTHER |
| 152 | 1007.5416 | 10.2 | [M + HCOO]− | C48H82O19 | 491 | O-162/O-162-146 | Majoroside F6 | OTHER |
| 153 | 1009.559 | 5.7 | [M + HCOO]− | C48H84O19 | 493 | O-162/O-162-146 | β-d-Glucopyranoside, (3β,6α,12β)-20-(β-d-glucopyranosyloxy)-3,12,25-trihydroxydammaran-6-yl 2-O-(6-deoxy-α-L-mannopyranosyl)- (ACI) | OTHER |
| 154 | 1025.5541 | 10.6 | [M + HCOO]− | C48H84O20 | 493 | O-162-162/O-162 | Vinaginsenoside R13 isomer | OTHER |
| 155 | 1025.5541 | 12.0 | [M + HCOO]− | C48H84O20 | 493 | O-162/O-162-162 | Vinaginsenoside R13 | OTHER |
| 156 | 1137.6053 | 21.9 | [M + HCOO]− | C54H92O22 | 443 | O-162-162/O-162-162 | Notoginsenoside I | OTHER |
| 157 | 1151.5848 | 22.0 | [M + HCOO]− | C54H90O23 | 457 | O-162-162/O-162-162 | 5,6-Didehydroginsenoside Rb1 | OTHER |
| 158 | 1167.5913 | 8.5 | [M + HCOO]− | C54H90O24 | 473 | O-162-162/O-162-162 | Notoginsenoside B | OTHER |
| 159 | 1169.5943 | 10.2 | [M + HCOO]− | C54H92024 | 475 | O-162-162/O-162-162 | Koryoginsenoside R2 | OTHER |
| 160 | 1169.5943 | 12.6 | [M + HCOO]− | C54H92O24 | 475 | O-162-162/O-162-162 | Notoginsenoside A | OTHER |
| 161 | 781.4734 | 41.3 | [M + HCOO]− | C41H68O11 | 441 | O-162-132 | Notoginsenoside ST11 isomer | OTHER |
| 162 | 843.4948 | 19.7 | [M + HCOO]− | C42H70O14 | 473 | O-162-162 | 11-Oxomogroside II A1 | OTHER |
| 163 | 973.5373 | 16.2 | [M + HCOO]− | C48H80O17 | 473 | O-162-146/O-146 | (3β,16β,22α)-28-[(6-Deoxy-α-L-mannopyranosyl)oxy]-16,22-dihydroxyolean-12-en-3-yl 6-deoxy-3-O-β-d-glucopyranosyl-α-L-mannopyranoside | OTHER |
| 164 | 973.5373 | 15.0 | [M + HCOO]− | C48H80O17 | 473 | O-162-146/O-146 | (3β,16β,22α)-28-[(6-Deoxy-α-l-mannopyranosyl)oxy]-16,22-dihydroxyolean-12-en-3-yl 6-deoxy-3-O-β-d-glucopyranosyl-α-l-mannopyranoside isomer | OTHER |
| 165 | 1009.5575 | 19.1 | [M + HCOO]− | C48H84O19 | 477 | O-162-162/O-162 | (3β,12β)-20-(β-d-Glucopyranosyloxy)-12,25-dihydroxydammaran-3-yl 2-O-β-d-glucopyranosyl-β-d-glucopyranoside | OTHER |
| 166 | 1025.5541 | 5.1 | [M + HCOO]− | C48H84O20 | 509 | O-162/O-162-146 | (3β,12β)-20-(β-d-Glucopyranosyloxy)-3,12,24,25-tetrahydroxydammaran-6-yl 2-O-(6-deoxy-α-l-β-d-mannopyranosyl)-β-d-glucopyranoside | OTHER |
| 167 | 803.4426 | 34.1 | [M + HCOO]− | C39H66O14 | 433 | O-162-162 | PQ-ginsenoside A ** | OTHER |
| 168 | 803.4426 | 36.0 | [M + HCOO]− | C39H66O14 | 433 | O-162-162 | PQ-ginsenoside A isomer ** | OTHER |
| 169 | 803.4426 | 19.3 | [M + HCOO]− | C39H66O14 | 449 | O-162-146 | PQ-ginsenoside B ** | OTHER |
| 170 | 785.4322 | 26.4 | [M + HCOO]− | C40H66O15 | 431 | O-162-146 | PQ-ginsenoside C ** | OTHER |
| 171 | 785.4322 | 39.0 | [M + HCOO]− | C40H66O15 | 415 | O-162-162 | PQ-ginsenoside D ** | OTHER |
| 172 | 827.4438 | 41.7 | [M + HCOO]− | C41H66O14 | 415 | O-162-162 | Acetyl-PQ-ginsenoside D ** | OTHER |
| 173 | 827.4438 | 41.3 | [M + HCOO]− | C41H66O14 | 415 | O-162-162 | Acetyl-PQ-ginsenoside D isomer I ** | OTHER |
| 174 | 827.4438 | 42.0 | [M + HCOO]− | C41H66O14 | 415 | O-162-162 | Acetyl-PQ-ginsenoside D isomer II ** | OTHER |
| 175 | 845.4536 | 40.0 | [M + HCOO]− | C41H68O15 | 433 | O-162-162 | Acetyl-PQ-ginsenoside A ** | OTHER |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, J.; Liu, F.; Ji, W.; Wang, X.; Li, L. Comprehensive Investigation of Ginsenosides in the Steamed Panax quinquefolius with Different Processing Conditions Using LC-MS. Molecules 2024, 29, 623. https://doi.org/10.3390/molecules29030623
Fan J, Liu F, Ji W, Wang X, Li L. Comprehensive Investigation of Ginsenosides in the Steamed Panax quinquefolius with Different Processing Conditions Using LC-MS. Molecules. 2024; 29(3):623. https://doi.org/10.3390/molecules29030623
Chicago/Turabian StyleFan, Jiali, Feng Liu, Wenhua Ji, Xiao Wang, and Lili Li. 2024. "Comprehensive Investigation of Ginsenosides in the Steamed Panax quinquefolius with Different Processing Conditions Using LC-MS" Molecules 29, no. 3: 623. https://doi.org/10.3390/molecules29030623
APA StyleFan, J., Liu, F., Ji, W., Wang, X., & Li, L. (2024). Comprehensive Investigation of Ginsenosides in the Steamed Panax quinquefolius with Different Processing Conditions Using LC-MS. Molecules, 29(3), 623. https://doi.org/10.3390/molecules29030623