Synthesis, Structure and Antibacterial Activity of Two Novel Coordination Polymers Based on N,N′-bis(4-carbozvlbenzvl)-4-aminotoluene and Heterocyclic Ligand against S. aureus
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structure Description of Compound 1
2.2. Structure Description of Compound 2
2.3. PXRD and Thermogravimetric Analysis (TGA)
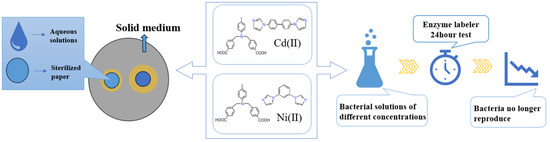
2.4. Culture of Bacteria and Fungi
2.5. Circle of Inhibition Test
2.6. Growth Curve Experiment
3. Experimental Section
3.1. Materials and Methods
3.2. Synthesis of [Cd2(L)2(bibp)2]n (1)
3.3. Synthesis of [Ni(L)(bib)]n (2)
3.4. X-ray Crystallography
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pettinari, C.; Pettinari, R.; Nicola, C.D.; Tombesi, A.; Scuri, S.; Marchetti, F. Antimicrobial MOFs. Coord. Chem. Rev. 2021, 446, 214121. [Google Scholar] [CrossRef]
- Liu, X.P.; Yan, Z.Q.; Zhang, Y.Y.; Zheng, W.L.; Sun, Y.H.; Ren, J.S.; Qu, X.G. Two-dimensional metal-organic framework/enzyme hybrid nanocatalyst as a benign and self-activated cascade reagent for in vivo wound healing. ACS Nano 2019, 13, 5222–5230. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.W.; Wang, S.H.; Xia, Y.Z.; Wu, L.; Huang, R.K.; He, Z.J. Frustrated Lewis pair boosting photocatalytic antibacterial activity on PDI-bridged bimetallic UiO-66-NH2. Dalton Trans. 2023, 52, 6813–6822. [Google Scholar] [CrossRef]
- Alavijeh, R.K.; Beheshti, S.; Akhbari, K.; Morsali, A. Investigation of Reasons for Metal-organic Framework’s Antibacterial Activities. Polyhedron 2018, 156, 257–278. [Google Scholar] [CrossRef]
- Quirós, J.; Boltes, K.; Aguado, S.; Villoria, R.G.D.; Vilatela, J.J.; Rosal, R. Antimicrobial metal-organic frameworks incorporated into electrospun fibers. Chem. Eng. J. 2015, 262, 189–197. [Google Scholar] [CrossRef]
- Brogan, D.M.; Mossialos, E. A Critical Analysis of the Review on Antimicrobial Resistance Report and the Infectious Disease Financing Facility. Glob. Health 2016, 12, 8. [Google Scholar] [CrossRef]
- Hemeg, H.A. Nanomaterials for alternative antibacterial therapy. Int. J. Nanomed. 2017, 12, 8211–8225. [Google Scholar] [CrossRef]
- Santo, C.E.; Quaranta, D.; Grass, G. Antimicrobial metallic copper surfaces kill Staphylococcus haemolyticus via membrane damage. MicrobiologyOpen 2012, 1, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, X.; Xia, Q.C.; Yuan, G.Z.; He, Q.Z.; Cui, H.Y. Multiple topological isomerism of three-connected networks in silver-based metal-organoboron frameworks. Chem. Commun. 2010, 46, 2608–2610. [Google Scholar] [CrossRef]
- Abbasi, A.R.; Akhbari, K.; Morsali, A. Dense coating of surface mounted CuBTCMetal-Organic Framework nanostructures on silk fibers, prepared by layer-by-layer methodunder ultrasound irradiation with antibacterial activity. Ultrason.-Sonochemistry 2012, 4, 846–852. [Google Scholar] [CrossRef]
- Restrepo, J.; Serroukh, Z.; Santiago-Morales, J.; Aguado, S.; Gomez-Sal, P.; Mosquera, M.E.G.; Rosal, R. An Antibacterial Zn-MOF with Hydrazinebenzoate Linkers. Berichte Der Dtsch. Chem. Ges. 2017, 3, 574–580. [Google Scholar] [CrossRef]
- . Khan, Z.A.; Goda, E.S.; Rehman, A.U.; Sohail, M. Selective antimicrobial and antibiofilm activity of metal–organic framework NH2-MIL-125 against Staphylococcus aureus. Mater. Sci. Eng. B. 2021, 269, 115146. [Google Scholar] [CrossRef]
- Ghaffar, I.; Imran, M.; Perveen, S.; Kanwal, T.; Saifullah, S.; Bertino, M.F.; Ehrhardt, C.J.; Yadavalli, V.K.; Shah, M.R. Synthesis of chitosan coated metal organic frameworks (MOFs) for increasing vancomycin bactericidal potentials against resistant S. aureus strain. Mater. Sci. Eng. C 2019, 105, 110111. [Google Scholar] [CrossRef]
- Skvortsova, A.; Kocianova, A.; Guselnikova, O.; Elashnikow, R.; Burtsev, V.; Rimpelova, S.; Pavlickova, V.S.; Fitl, P.; Lukesova, M.; Kolska, Z.; et al. Self-activated antibacterial MOF-based coating on medically relevant polypropylene. Appl. Surf. Sci. 2023, 623, 157048. [Google Scholar] [CrossRef]
- Zhang, S.Q.; Ye, J.W.; Sun, Y.; Kang, J.; Liu, J.H.; Wang, Y.; Li, Y.C.; Zhang, L.H.; Ning, G.L. Electrospun fibrous mat based on silver (I) metal-organic frameworkspolylactic acid for bacterial killing and antibiotic-free wound dressing. Chem. Eng. J. 2020, 390, 124523. [Google Scholar] [CrossRef]
- Hu, W.C.; Pang, J.; Biswas, S.; Wang, K.; Wang, C.; Xia, X.H. Ultrasensitive Detection of Bacteria Using a 2D MOF Nanozyme Amplified Electrochemical Detector. Anal. Chem. 2021, 93, 8544–8552. [Google Scholar] [CrossRef]
- Li, X.Q.; Zhao, X.S.; Chu, D.D.; Zhu, X.L.; Xue, B.L.; Chen, H.; Zhou, Z.; Li, J.G. Silver nanoparticle-decorated 2D Co-TCPP MOF nanosheets for synergistic photodynamic and silver ion antibacterial. Surf. Interfaces 2022, 33, 102247. [Google Scholar] [CrossRef]
- Lu, X.Y.; Ye, J.W.; Zhang, D.K.; Xie, R.X.; Bogale, R.F.; Sun, Y.; Zhao, L.M.; Zhao, Q.; Ning, G.L. Silver carboxylate metal–organic frameworks with highly antibacterial activity and biocompatibility. Inorg. Biochem. 2014, 138, 114–121. [Google Scholar] [CrossRef]
- Liu, Z.; Ye, J.W.; Rauf, A.; Zhang, S.Q.; Wang, G.Y.; Shi, S.Q.; Ning, G.L. A flexible fibrous membrane based on copper(ii) metal–organic framework/poly(lactic acid) composites with superior antibacterial performance. Biomater. Sci. 2021, 9, 3851–3859. [Google Scholar] [CrossRef]
- Xiong, F.F.; Zhang, T.; Ma, T.; Jia, Q. Methods to characterize and discover molecular degraders in cells. Talanta 2024, 266, 125068. [Google Scholar] [CrossRef]
- Li, W.; Lin, J.; Zhou, J.; He, S.Q.; Wang, A.Q.; Hu, Y.F.; Li, H.M.; Zou, L.; Liu, Y. Hyaluronic acid-functionalized DDAB/PLGA nanoparticles for improved oral delivery of magnolol in the treatment of ulcerative colitis. Int. J. Pharm. 2024, 653, 123878. [Google Scholar] [CrossRef] [PubMed]










| Compound | 1 | 2 |
|---|---|---|
| CCDC No. | 2338641 | 2338643 |
| Molecular Formula | C82H66N10O8Cd2 | C35H29N5O4Ni |
| Fw | 1544.27 | 642.32 |
| Crystal system | Triclinic | Orthorhombic |
| Space group | P1 | Fdd2 |
| a/Å | 10.9792(7) | 19.6165(14) |
| b/Å | 11.6582(8) | 54.500(4) |
| c/Å | 13.7423(9) | 11.1375(8) |
| α/(°) | 81.8140(10) | 90 |
| β/(°) | 86.0920(10) | 90 |
| γ/(°) | 88.6630(10) | 90 |
| V/Å3 | 1736.8(2) | 11907.2(15) |
| Z | 1 | 16 |
| Dc/(g⋅cm−3) | 1.897 | 1.615 |
| F (000) | 920 | 5760 |
| GOF on F2 | 1.038 | 1.037 |
| R1/wR2 [I > 2σ(I)] | 0.0347, 0.0756 | 0.0574, 0.1009 |
| R1/wR2 (all data) | 0.0496, 0.0819 | 0.1251, 0.1319 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Zhao, L.; Jing, H.; Song, G.; Li, J. Synthesis, Structure and Antibacterial Activity of Two Novel Coordination Polymers Based on N,N′-bis(4-carbozvlbenzvl)-4-aminotoluene and Heterocyclic Ligand against S. aureus. Molecules 2024, 29, 1990. https://doi.org/10.3390/molecules29091990
Wang Z, Zhao L, Jing H, Song G, Li J. Synthesis, Structure and Antibacterial Activity of Two Novel Coordination Polymers Based on N,N′-bis(4-carbozvlbenzvl)-4-aminotoluene and Heterocyclic Ligand against S. aureus. Molecules. 2024; 29(9):1990. https://doi.org/10.3390/molecules29091990
Chicago/Turabian StyleWang, Ziyun, Lun Zhao, Hongwei Jing, Guanying Song, and Jiayu Li. 2024. "Synthesis, Structure and Antibacterial Activity of Two Novel Coordination Polymers Based on N,N′-bis(4-carbozvlbenzvl)-4-aminotoluene and Heterocyclic Ligand against S. aureus" Molecules 29, no. 9: 1990. https://doi.org/10.3390/molecules29091990
APA StyleWang, Z., Zhao, L., Jing, H., Song, G., & Li, J. (2024). Synthesis, Structure and Antibacterial Activity of Two Novel Coordination Polymers Based on N,N′-bis(4-carbozvlbenzvl)-4-aminotoluene and Heterocyclic Ligand against S. aureus. Molecules, 29(9), 1990. https://doi.org/10.3390/molecules29091990