Genetic and Association Mapping Study of Wheat Agronomic Traits Under Contrasting Water Regimes
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Evaluation
2.2. Heritability of the Traits
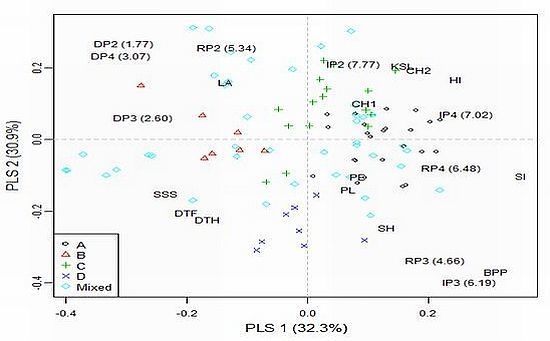
2.3. Genetic Correlations
2.4. Model-Based Groups within AM Population and Linkage Disequilibrium
2.5. Marker-Trait Association
3. Discussion
4. Experimental Section
4.1. Plant Material
4.2. Field Trials and Experimental Data
4.3. Genotype Analysis
4.4. Phenotypic Data Analysis
4.5. Population Structure
4.6. Linkage Disequilibrium
4.7. Association Analysis
5. Conclusion
Supplementary Materials
ijms-13-06167-s001.pdfAcknowledgments
References
- IPPC, Working Group II: “Impacts, Adaptation and Vulnerability”. In Fourth Assessment Report; Intergovernmental Panel on Climate Change; Stanford, CA, USA, 2007.
- Reynolds, M.P.; Mujeeb-Kazi, A.; Sawkins, M. Prospects of utilizing plant adaptive mechanisms to improve wheat and other crops in drought and salinity prone environments. Ann. Appl. Biol 2005, 146, 239–259. [Google Scholar]
- Räisänen, J.; Hansson, U.; Ullerstig, A.; Döscher, R.; Graham, L.P.; Jones, C.; Meier, H.E.M.; Samuelsson, P.; Willén, U. European climate in the late 21st century: Regional simulations with two driving global models and two forcing scenarios. Clim. Dyn 2004, 22, 13–31. [Google Scholar]
- Döll, P.; Flörke, M. Global-scale Estimation of Diffuse Groundwater Recharge. In Frankfurt Hydrology Paper 03; Institute of Physical Geography, Frankfurt University: Frankfurt am Main, Germany, 2005. [Google Scholar]
- IPCC. Working Group II: “Impacts, Adaptation and Vulnerability”. Third Assessment Report; Intergovernmental Panel on Climate Change, Stanford, CA, USA; 2001.
- Salekdeh, G.S.; Reynolds, M.; Bennett, J.; Boyer, J. Conceptual framework for drought phenotyping during molecular breeding. Trends Plant Sci 2009, 14, 488–496. [Google Scholar]
- Stich, B.; Melchinger, A.E. An introduction to association mapping in plants. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour 2010, 5, 1–9. [Google Scholar]
- Quarrie, S.A.; Steed, A.; Calestani, C.; Semikhodskii, A.; Lebreton, C.; Steele, N.; Pljevljakusić, D.; Waterman, E.; Weyen, J.; et al. A high density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring × SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor. Appl. Genet 2005, 110, 865–880. [Google Scholar]
- Crossa, J.; Burgueño, J.; Dreisigacker, S.; Vargas, M.; Herrera-Foessel, S.A.; Lillemo, M.; Singh, R.P.; Trethowan, R.; Warburton, M.; Franco, J.; et al. Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics 2007, 177, 1889–1913. [Google Scholar]
- McIntyre, C.L.; Mathews, K.L.; Rattey, A.; Chapman, S.C.; Drenth, J.; Ghaderi, M.; Reynolds, M.; Shorter, R. Molecular detection of genomic regions associated with grain yield and yield-related components in an elite bread wheat cross evaluated under irrigated and rainfed conditions. Theor. Appl. Genet 2010, 120, 527–541. [Google Scholar]
- Maccaferri, M.; Sanguineti, M.C.; Demontis, A.; El-Ahmed, A.; del Moral, L.G.; Maalouf, F.; Nachit, M.; Nserallah, N.; Ouabbou, H.; Rhouma, S.; et al. Association mapping in durum wheat grown across a broad range of water regimes. J. Exp. Bot 2011, 62, 409–438. [Google Scholar]
- Breseghello, F.; Sorrells, M.E. Association analysis as a strategy for improvement of quantitative traits in plants. Crop Sci 2006, 46, 1323–1330. [Google Scholar]
- Neumann, K.; Kobiljski, B.; Denčić, S.; Varshney, R.K.; Börner, A. Genome-wide association mapping: A case study in bread wheat (Triticum aestivum L.). Mol. Breed 2011, 27, 37–58. [Google Scholar]
- Yang, D.L.; Jing, R.L.; Chang, X.P.; Li, W. Identification of quantitative trait loci and environmental interactions for accumulation and remobilization of water-soluble carbohydrates in wheat (Triticum aestivum L.) stems. Genetics 2007, 176, 571–584. [Google Scholar]
- Yang, D.L.; Jing, R.L.; Chang, X.P.; Li, W. Quantitative trait loci mapping for chlorophyll fluorescence and associated traits in wheat (Triticum aestivum). J. Integr. Plant Biol 2007, 49, 646–654. [Google Scholar]
- Rebetzke, G.J.; Condon, A.G.; Farquhar, G.D.; Appels, R.; Richards, R.A. Quantitative trait loci for carbon isotope discrimination are repeatable across environments and wheat mapping populations. Theor. Appl. Genet 2008, 118, 123–137. [Google Scholar]
- Rebetzke, G.J.; van Herwaarden, A.F.; Jenkins, C.; Weiss, M.; Lewis, D.; Ruuska, S.; Tabe, L.; Fettell, N.A.; Richards, R.A. Quantitative trait loci for soluble stem carbohydrate production in wheat. Aust. J. Agric. Res 2008, 59, 891–905. [Google Scholar]
- Wang, R.; Yu, Y.; Zhao, J.; Shi, Y.; Song, Y.; Wang, T.; Li, Y. Population structure and linkage disequilibrium of a mini core set of maize inbred lines in China. Theor. Appl. Genet 2008, 117, 1141–1153. [Google Scholar]
- Whitt, S.R.; Buckler, E.S. Using natural allelic diversity to evaluate gene function. Methods Mol. Biol 2003, 236, 123–139. [Google Scholar]
- Kobiljski, B.; Quarrie, S.A.; Denčić, S.; Kirby, J.; Ivegeš, M. Genetic diversity of the Novi Sad wheat core collection revealed by microsatellites. Cell Mol. Biol. Lett 2002, 7, 685–694. [Google Scholar]
- Dodig, D.; Zorić, M.; Kobiljski, B.; Šurlan-Momirović, G.; Quarrie, S.A. Assessing drought tolerance and regional patterns of genetic diversity among spring and winter bread wheat using simple sequence repeats and phenotypic data. Crop Pasture Sci 2010, 61, 812–824. [Google Scholar]
- Somers, D.J.; Banks, T.; DePauw, R.; Fox, S.; Clarke, J.; Pozniak, C.; McCartney, C. Genome-wide linkage disequilibrium analysis in bread wheat and durum wheat. Genome 2007, 50, 557–567. [Google Scholar]
- Chao, S.; Zhang, V.; Dubcovsky, J.; Sorrells, M.E. Evaluation of genetic diversity and genome-wide linkage disequilibrium among US wheat (Triticum aestivum L.) germplasm representing different marker classes. Crop Sci 2007, 47, 1018–1030. [Google Scholar]
- Prasad, B.; Babar, M.A.; Xu, X.Y.; Bai, G.H.; Klatt, A.R. Genetic diversity in U.S. hard red winter wheat cultivars as revealed by microsatellite markers. Crop Pasture Sci 2009, 60, 16–24. [Google Scholar]
- Cooper, M.; Woodruff, D.R.; Eisemann, R.L.; Brennan, P.S.; DeLacy, I.H. A selection strategy to accommodate genotype-by-environment interaction for grain yield of wheat: Managed-environments for selection among genotypes. Theor. Appl. Genet 1995, 90, 492–502. [Google Scholar]
- Trethowan, R.M.; van Ginkel, M.; Rajaram, S. Progress in breeding for yield and adaptation in global drought affected environments. Crop Sci 2002, 42, 1441–1446. [Google Scholar]
- McCartney, C.A.; Somers, D.J.; Humphreys, D.G.; Lukow, O.; Ames, N.; Noll, J.; Cloutier, S.; McCallum, B.D. Mapping quantitative trait loci controlling agronomic traits in the spring wheat cross RL4452 X ‘AC Domain’. Genome 2005, 48, 870–883. [Google Scholar]
- Golabadi, M.; Arzani, A.; Mirmohammadi Maibody, S.A.M.; Sayed Tabatabaei, B.E.; Mohammadi, S.A. Identification of microsatellite markers linked with yield components under drought stress at terminal growth stages in durum wheat. Euphytica 2011, 177, 207–221. [Google Scholar]
- Remington, D.L.; Thornsberry, J.M.; Matsuoka, Y.; Wilson, L.M.; Whitt, S.R.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 2001, 98, 11479–11484. [Google Scholar]
- Abdurakhmonov, I.Y.; Kohel, R.J.; Yu, J.Z.; Pepper, A.E.; Abdullaev, A.A.; Kushanov, F.N.; Salakhutdinov, I.B.; Buriev, Z.T.; Saha, S.; Scheffler, B.E.; et al. Molecular diversity and association mapping of fiber quality traits in exotic G. hirstum L. germplasm. Genomics 2008, 92, 478–487. [Google Scholar]
- Malysheva-Otto, L.V.; Ganal, M.W.; Roder, M.S. Analysis of molecular diversity, population structure and linkage disequilibrium in a worldwide survey of cultivated barley germplasm (Hordeum vulgare L.). BMC Genet 2006, 7. [Google Scholar] [CrossRef]
- Maccaferri, M.; Sanguineti, M.C.; Donini, P.; Tuberosa, R. Population structure and long range linkage disequilibrium in a durum wheat elite collection. Mol. Breed 2005, 15, 271–289. [Google Scholar]
- Slafer, G.A. Genetic basis of yield as viewed from a crop physiologist’s perspective. Ann. Appl. Biol 2003, 142, 117–128. [Google Scholar]
- Zhang, L.Y.; Liu, D.C.; Guo, X.L.; Yang, W.L.; Sun, J.Z.; Wang, D.W.; Zhang, A.M. Genomic distribution of quantitative trait loci for yield and yield related traits in common wheat. J. Int. Plant Biol 2010, 52, 996–1007. [Google Scholar]
- Somers, D.J.; Isaac, P.; Edwards, K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L). Theor. Appl. Genet 2004, 109, 1105–1114. [Google Scholar]
- Kumar, N.; Kulwal, P.L.; Balyan, H.S.; Gupta, P.K. QTL mapping for yield and yield contribution traits in two mapping populations of bread wheat. Mol. Breed 2007, 19, 163–177. [Google Scholar]
- Hanocq, E.; Niarquin, M.; Heumez, E.; Rousset, M.; le Gouis, J. Detection and mapping of QTL for earliness components in a bread wheat recombinant inbred lines population. Theor. Appl. Genet 2004, 110, 106–115. [Google Scholar]
- Worland, A.J. The influence of flowering time genes on environmental adaptability in European wheats. Euphytica 1996, 89, 49–57. [Google Scholar]
- Guo, Z.; Song, Y.; Zhou, R.; Ren, Z.; Jia, J. Discovery, evaluation and distribution of haplotypes of the wheat Ppd-D1 gene. New Phytol 2010, 185, 841–851. [Google Scholar]
- Ruuska, S.A.; Rebetzke, G.J.; van Herwaarden, A.F.; Richards, R.A.; Fettell, N.A.; Tabe, L.; Jenkins, C.L.D. Genotypic variation in water-soluble carbohydrate accumulation in wheat. Funct. Plant Biol 2006, 33, 799–809. [Google Scholar]
- Ehdaie, B.; Alloush, G.A.; Waines, J.G. Genotypic variation in linear rate of grain growth and contribution of stem reserves to grain yield in wheat. Field Crops Res 2008, 106, 34–43. [Google Scholar]
- Dreccer, M.F.; van Herwaarden, A.F.; Chapman, S.C. Grain number and grain weight in wheat lines contrasting for stem water soluble carbohydrate concentration. Field Crops Res 2009, 112, 43–54. [Google Scholar]
- Bezant, J.; Laurie, D.; Pratchett, N.; Chojecki, J.; Kearsey, M. Mapping QTL controlling yield and yield components in a spring barley (Hordeum vulgare L) cross using marker regression. Mol. Breed 1997, 29, 29–38. [Google Scholar]
- Marza, F.; Bai, G.H.; Carver, B.F.; Zhou, W.C. Quantitative trait loci for yield and related traits in the wheat population Ning7840 × Clark. Theor. Appl. Genet 2006, 112, 688–698. [Google Scholar]
- Kirigwi, F.M.; van Ginkel, M.; Brown-Guedira, G.; Gill, B.S.; Paulsen, G.M.; Fritz, A.K. Markers associated with a QTL for grain yield in wheat under drought. Mol. Breed 2007, 20, 401–413. [Google Scholar]
- Quarrie, S.A.; Pekić-Quarrie, S.; Radošević, R.; Rančić, D.; Dodig, D. Dissecting a wheat QTL for yield present in a range of environments: From the QTL to candidate genes. J. Exp. Bot 2006, 57, 2627–2637. [Google Scholar]
- Quarrie, S.A.; Dodig, D.; Pekić, S.; Kirby, J.; Kobiljski, B. Prospects for marker-assisted selection of improved drought responses in wheat. Bulg. J. Plant Physiol 2003, 83–95. [Google Scholar]
- Peleg, Z.; Fahima, T.; Krugman, T.; Abbo, S.; Yakir, D.; Korol, A.B.; Saranga, Y. Genomic dissections of drought resistance in durum wheat × wild emmer wheat recombinant inbreed line population. Plant. Cell Environ 2009, 32, 758–779. [Google Scholar]
- Dashti, H.; Yazdi-Samadi, B.; Ghannadha, M.; Naghavi, M.R.; Quarrie, S. QTL analysis for drought resistance in wheat using doubled haploid lines. Int. J. Agric. Biol 2007, 9, 98–101. [Google Scholar]
- Kobiljski, B.; Denčić, S.; Kondić-Špika, A.; Lohwasser, U.; Börner, A. Locating stable across environment QTL involved in the determination of agronomic characters in wheat. Cereal Res. Commun 2009, 37, 327–333. [Google Scholar]
- Dodig, D.; Quarrie, S.A.; Stanković, S.; Milijić, S.; Denčić, S. Characterising Wheat Genetic Resources for Responses to Drought Stress [CD-ROM]. Proceedings of the International Conference on Drought Mitigation and Prevention of Land Desertification, Bled, Slovenia, 21–25 April 2002.
- Fischer, R.A.; Maurer, R. Drought resistance in spring wheat cultivars: I. Grain yield responses. Aust. J. Agric. Res 1978, 29, 897–912. [Google Scholar]
- Rosielle, A.A.; Hamblin, J. Theoretical aspects of selection for yield in stress and non-stress environments. Crop Sci 1981, 21, 943–946. [Google Scholar]
- Hopwood, A.; Oldroyd, N.; Fellows, S.; Ward, R.; Owen, S.A.; Sullivan, K. Rapid quantification of DNA samples extracted from buccal scrapes prior to DNA profiling. Biotechniques 1997, 23, 18–20. [Google Scholar]
- Röder, M.S.; Korzun, V.; Wendehake, K.; Plaschke, J.; Tixier, M.H.; Leroy, P.; Ganal, M.W. A microsatellite map of wheat. Genetics 1998, 149, 2007–2023. [Google Scholar]
- Holland, J.B.; Nyquist, W.E.; Cervantes-Martinez, C.T. Estimating and interpreting heritability for plant breeding: An update. Plant Breed. Rev 2003, 22, 9–112. [Google Scholar]
- Holland, J.B. Estimating genotypic correlations and their standard errors using multivariate restricted maximum likelihood estimation with SAS Proc MIXED. Crop Sci 2006, 46, 642–654. [Google Scholar]
- Aastveit, A.; Martens, H. ANOVA interaction interpreted by partial least squares regression. Biometrics 1986, 42, 829–844. [Google Scholar]
- Vargas, M.; Crossa, J.; Sayre, K.; Reynolds, M.; Ramirez, M.E.; Talbot, M. Interpreting genotype × environment interaction in wheat using partial least squares regression. Crop Sci 1998, 38, 679–689. [Google Scholar]
- The R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008. Available online: http://www.R-project.org accessed on 13 November 2011.
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Falush, D.; Stephens, M.; Prithchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar]
- Witt, S.R.; Buckler, E.S. Using natural allelic diversity to evaluate gene function. Methods Mol. Biol 2003, 236, 123–139. [Google Scholar]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar]

| Trait | Trait Code † | Means ± SEM | DP/IP | ||
|---|---|---|---|---|---|
| IP | RP | DP | |||
| Days to heading | DTH(3) | 138.9 ± 0.40 a | 138.4 ± 0.41 a | 134.4 ± 0.45 b | 96.8 |
| Days to flowering | DTF(3) | 144.2 ± 0.54 a | 143.5 ± 0.54 a | 139.1 ± 0.56 b | 96.5 |
| Heading-flowering (days) | HF(3) | 5.46 ± 0.95 a | 5.31 ± 0.10 a,b | 4.96 ± 0.11 b | 90.8 |
| Stem height (cm) | SH(3) | 71.3 ± 1.32 a | 64.4 ± 1.25 b | 65.2 ± 1.28 a,b | 91.4 |
| Peduncle length (cm) | PL(2) | 30.0 ± 0.70 a | 28.0 ± 0.72 b | 30.1 ± 0.60 a | 100.3 |
| Peduncle extrusion (cm) | PE(2) | 11.6 ± 0.55 a,b | 10.6 ± 0.56 b | 13.0 ± 0.46 a | 112.1 |
| Prod. tillering (No/plant) | PT(3) | 2.45 ± 0.04 a | 2.10 ± 0.03 b | 1.28 ± 0.02 c | 52.2 |
| Spike length (cm) | SL(3) | 7.91 ± 0.10 a | 7.62 ± 0.10 a | 6.99 ± 0.09 b | 88.4 |
| Spike density | SD(3) | 2.12 ± 0.02 a | 2.13 ± 0.03 a | 2.15 ± 0.03 a | 101.4 |
| Fertile spikelets per spike | FSS(3) | 14.9 ± 0.15 a | 14.2 ± 0.14 b | 12.1 ± 0.10 c | 81.2 |
| Sterile spikelets per spike | SSS(3) | 1.58 ± 0.09 b | 1.77 ± 0.10 b | 2.56 ± 0.11 a | 162.0 |
| Kernels per spike | KS(3) | 38.5 ± 0.54 a | 36.5 ± 0.51 a | 28.1 ± 0.38 b | 73.0 |
| Kernels per spikelets | KSL(3) | 2.36 ± 0.04 a | 2.32 ± 0.04 a | 1.99 ± 0.03 b | 81.8 |
| Kernel number (m−2) | KN(3) | 18801 ± 272 a | 15202 ± 238 b | 7369 ± 143 c | 39.2 |
| 1000 kernel weight (g) | TKW(3) | 37.1 ± 0.37 a | 35.8 ± 0.36 a | 33.5 ± 0.33 b | 90.3 |
| Biomass per plant (g) | BPP(3) | 7.69 ± 0.11 a | 6.19 ± 0.09 b | 2.90 ± 0.05 c | 37.7 |
| Harvest index | HI(3) | 0.44 ± 0.01 a | 0.43 ± 0.01 a | 0.41 ± 0.00 b | 93.2 |
| Production per spike (g) | PPS(3) | 1.43 ± 0.03 a | 1.31 ± 0.02 b | 0.94 ± 0.02 c | 65.7 |
| Spike index | SI(3) | 0.73 ± 0.00 a | 0.72 ± 0.00 a | 0.72 ± 0.00 a | 98.6 |
| Leaf area (cm−2) | LA(2) | 29.0 ± 0.50 a | – | 24.0 ± 0.45 b | 82.8 |
| Leaf width (mm) | LW(2) | 22.1 ± 0.20 a | – | 20.7 ± 0.22 b | 93.7 |
| Chlorophyll at flowering †† | CH1(2) | 51.0 ± 0.28 a | – | 51.7 ± 0.29 a | 101.4 |
| Chlorophyll at gr. filling †† | CH2(2) | 32.1 ± 0.58 a | – | 30.3 ± 0.71 b | 94.4 |
| Grain yield (t·ha−1) | GY(3) | 6.99 ± 0.12 a | 5.49 ± 0.11 b | 2.48 ± 0.05 c | 35.5 |
| Trait | IP | RP | DP | Overall |
|---|---|---|---|---|
| DTH | 0.90 | 0.90 | 0.85 | 0.90 |
| DTF | 0.88 | 0.87 | 0.84 | 0.88 |
| HF | 0.46 | 0.48 | 0.26 | 0.37 |
| SH | 0.92 | 0.86 | 0.83 | 0.89 |
| PL | 0.80 | 0.91 | 0.89 | 0.87 |
| PE | 0.74 | 0.87 | 0.82 | 0.82 |
| PT | 0.66 | 0.69 | 0.52 | 0.52 |
| SL | 0.93 | 0.93 | 0.88 | 0.91 |
| SD | 0.88 | 0.88 | 0.89 | 0.89 |
| FSS | 0.83 | 0.82 | 0.77 | 0.78 |
| SSS | 0.84 | 0.87 | 0.83 | 0.85 |
| KS | 0.79 | 0.82 | 0.78 | 0.79 |
| KSL | 0.68 | 0.73 | 0.70 | 0.73 |
| KN | 0.61 | 0.64 | 0.38 | 0.48 |
| TKW | 0.87 | 0.84 | 0.71 | 0.81 |
| BPP | 0.64 | 0.57 | 0.39 | 0.44 |
| HI | 0.82 | 0.80 | 0.70 | 0.80 |
| PPS | 0.71 | 0.76 | 0.77 | 0.78 |
| SI | 0.72 | 0.55 | 0.66 | 0.64 |
| LA | 0.78 | - | 0.76 | 0.79 |
| LW | 0.72 | - | 0.78 | 0.77 |
| CH1 | 0.75 | - | 0.70 | 0.71 |
| CH2 | 0.66 | - | 0.58 | 0.57 |
| GY | 0.74 | 0.75 | 0.47 | 0.55 |
| Trait | IP | RP | DP | Overall |
|---|---|---|---|---|
| DTH | −0.30 ** | −0.28 * | −0.08 | −0.25 * |
| DTF | −0.36 ** | −0.35 ** | −0.11 | −0.31 ** |
| HF | −0.20 | −0.25 * | −0.25 * | −0.22 * |
| SH | 0.22 * | 0.27 ** | 0.31 ** | 0.27 ** |
| PL | 0.21 * | 0.18 | 0.12 | 0.16 |
| PE | 0.22 * | 0.14 | 0.07 | 0.15 |
| PT | 0.36 ** | 0.35 ** | 0.33 ** | 0.40 ** |
| SL | −0.11 | −0.08 | 0.11 | −0.05 |
| SD | −0.07 | −0.11 | −0.15 | −0.12 |
| FSS | 0.02 | 0.07 | 0.33 ** | 0.11 |
| SSS | −0.42 ** | −0.41 ** | −0.31 ** | −0.42 ** |
| KS | 0.16 | 0.23 * | 0.29 ** | 0.21 * |
| KSL | 0.35 ** | 0.40 ** | 0.27 ** | 0.34 ** |
| KN | 0.82 ** | 0.86 ** | 0.81 ** | 0.83 ** |
| TKW | 0.64 ** | 0.63 ** | 0.57 ** | 0.61 ** |
| BPP | 0.75 ** | 0.82 ** | 0.86 ** | 0.79 ** |
| HI | 0.51 ** | 0.52 ** | 0.51 ** | 0.54 ** |
| PPS | 0.43 ** | 0.47 ** | 0.56 ** | 0.47 ** |
| SI | 0.66 ** | 0.80 ** | 0.65 ** | 0.75 ** |
| LA | −0.20 | - | −0.40 ** | −0.13 |
| LW | −0.04 | - | −0.16 | 0.04 |
| CH1 | 0.30 ** | - | 0.34 ** | 0.30 ** |
| CH2 | 0.40 ** | - | 0.31 ** | 0.39 ** |
| DSI | 0.36 ** | 0.30 ** | −0.16 | 0.25 ** |
| TOL | 0.97 ** | 0.95 ** | 0.72 ** | 0.94 ** |
| Trait | Phenotype-Population Structure Associations | |
|---|---|---|
| R2 (%) a | P-Value a | |
| DTH | 6.0 | 0.034 |
| DTF | 5.7 | 0.039 |
| HF | 0.0 | - |
| SH | 57.8 | 0.000 |
| PL | 53.3 | 0.000 |
| PE | 42.9 | 0.000 |
| PT | 3.6 | 0.094 |
| SL | 0.0 | - |
| SD | 3.1 | 0.118 |
| FSS | 5.0 | 0.054 |
| SSS | 12.6 | 0.002 |
| KS | 2.4 | 0.156 |
| KSL | 6.0 | 0.034 |
| KN | 3.6 | 0.095 |
| TKW | 1.2 | 0.253 |
| BPP | 25.6 | 0.000 |
| HI | 19.3 | 0.000 |
| PPS | 3.9 | 0.084 |
| SI | 9.1 | 0.008 |
| LA | 6.1 | 0.032 |
| LW | 3.0 | 0.120 |
| CH1 | 12.8 | 0.001 |
| CH2 | 12.7 | 0.001 |
| GY | 6.4 | 0.028 |
| DSI | 0.0 | - |
| TOL | 2.1 | 0.179 |
| Trait | Marker | Chromosome Arm | R2a,b | MTA Consistancy d | |||
|---|---|---|---|---|---|---|---|
| IP | RP | DP | Overall c | ||||
| DTH | gwm257 | 2BS | 0.137 | 0.136 | 0.132 | 0.134 | 6 |
| DTH | psp3200 | 6DS | 0.144 | 0.181 | 0.192 | 0.165 | 6 |
| DTF | gwm257 | 2BS | 0.129 | 0.143 | 0.125 | 0.136 | 5 |
| DTF | gwm484 | 2DS | 0.197 | 0.229 | 0.252 | 0.228 | 5 |
| DTF | psp3200 | 6DS | 0.141 | 0.172 | 0.181 | 0.175 | 6 |
| PT | gwm484 | 2DS | 0.311 | 0.253 | 0.293 | 0.288 | 6 |
| PT | gwm389 | 3BS | NS | NS | 0.144 | NS | 2 |
| PT | psp3200 | 6DS | 0.139 | 0.149 | 0.160 | 0.161 | 6 |
| SL | gwm484 | 2DS | 0.341 | 0.356 | 0.361 | 0.360 | 8 |
| SL | psp3200 | 6DS | 0.271 | 0.257 | 0.288 | 0.273 | 9 |
| FSS | gwm484 | 2DS | 0.422 | 0.420 | 0.401 | 0.414 | 7 |
| FSS | psp3200 | 6DS | 0.330 | 0.318 | 0.302 | 0.324 | 9 |
| SSS | gwm484 | 2DS | 0.328 | 0.313 | 0.285 | 0.322 | 6 |
| SSS | gwm369 | 3AS | 0.208 | NS | NS | NS | 4 |
| KS | gwm540 | 5BS | 0.253 | 0.199 | NS | 0.238 | 4 |
| KS | psp3200 | 6DS | 0.284 | 0.252 | 0.266 | 0.262 | 7 |
| KN | psp3088 | 2AL | 0.138 | NS | 0.127 | 0.132 | 4 |
| HI | gwm458 | 1DL | NS | NS | 0.122 | NS | 2 |
| HI | gwm484 | 2DS | 0.217 | 0.202 | NS | 0.202 | 4 |
| PPS | psp3200 | 6DS | 0.127 | 0.152 | 0.167 | 0.148 | 7 |
| PPS | gwm130 | 7AS | NS | NS | 0.165 | 0.153 | 3 |
| LA | gwm484 | 2DS | 0.220 | ND | 0.280 | 0.240 | 4 |
| LA | psp3200 | 6DS | 0.232 | ND | 0.257 | 0.250 | 4 |
| LW | gwm484 | 2DS | NS | ND | 0.289 | 0.258 | 2 |
| LW | psp3050 | 7AS | NS | ND | 0.138 | 0.131 | 2 |
| LW | psp3200 | 6DS | 0.230 | ND | 0.244 | 0.243 | 4 |
| CH2 | gwm484 | 2DS | 0.213 | ND | 0.291 | 0.259 | 4 |
| GY | gwm99 | 1AL | 0.144 | NS | NS | NS | 2 |
| GY | gwm484 | 2DS | 0.223 | NS | NS | 0.198 | 3 |
| DSI | gwm484 | 2DS | NA | NA | NA | 0.253 | 2e |
| TOL | gwm257 | 2BS | NA | NA | NA | 0.149 | 2e |
| TOL | gwm484 | 2DS | NA | NA | NA | 0.228 | 2e |
| Season | Nov.–Feb. | March–June | Date of Shelter Installation | ||
|---|---|---|---|---|---|
| Drought Plots | Rainfed Plots | Irrigated Plots a | |||
| 2001/02 | 54.3 | 0.0 | 154.3 | +180 | 28 February |
| 2002/03 | 158.3 | 5.8 | 200.6 | +50 | 12 March |
| 2003/04 | 195.3 | 3.5 | 202.9 | +20 | 8 March |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Dodig, D.; Zoric, M.; Kobiljski, B.; Savic, J.; Kandic, V.; Quarrie, S.; Barnes, J. Genetic and Association Mapping Study of Wheat Agronomic Traits Under Contrasting Water Regimes. Int. J. Mol. Sci. 2012, 13, 6167-6188. https://doi.org/10.3390/ijms13056167
Dodig D, Zoric M, Kobiljski B, Savic J, Kandic V, Quarrie S, Barnes J. Genetic and Association Mapping Study of Wheat Agronomic Traits Under Contrasting Water Regimes. International Journal of Molecular Sciences. 2012; 13(5):6167-6188. https://doi.org/10.3390/ijms13056167
Chicago/Turabian StyleDodig, Dejan, Miroslav Zoric, Borislav Kobiljski, Jasna Savic, Vesna Kandic, Steve Quarrie, and Jeremy Barnes. 2012. "Genetic and Association Mapping Study of Wheat Agronomic Traits Under Contrasting Water Regimes" International Journal of Molecular Sciences 13, no. 5: 6167-6188. https://doi.org/10.3390/ijms13056167
APA StyleDodig, D., Zoric, M., Kobiljski, B., Savic, J., Kandic, V., Quarrie, S., & Barnes, J. (2012). Genetic and Association Mapping Study of Wheat Agronomic Traits Under Contrasting Water Regimes. International Journal of Molecular Sciences, 13(5), 6167-6188. https://doi.org/10.3390/ijms13056167