Recent Advances in Understanding the Control of Secretory Proteins by the Unfolded Protein Response in Plants
Abstract
:1. Introduction
2. Occurrence of ER Stress
3. Signaling Components
3.1. ER Stress Sensors
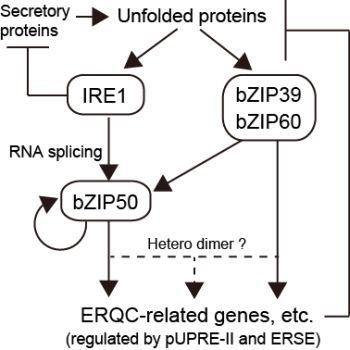
3.1.1. IRE1
3.1.2. ATF6-Like Transcription Factors
3.2. Transcription Factors Regulated by IRE1-Mediated Unconventional mRNA Splicing
3.3. cis-Elements
4. Upregulation of Gene Expression
5. Downregulation of Gene Expression
6. Interaction with Other Life Phenomena
7. Conclusions
Acknowledgments
Conflict of Interest
References
- Schubert, U.; Antón, L.C.; Gibbs, J.; Norbury, C.C.; Yewdell, J.W.; Bennink, J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 2000, 404, 770–774. [Google Scholar]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086. [Google Scholar]
- Mori, K. Signalling pathways in the unfolded protein response: Development from yeast to mammals. J. Biochem 2009, 146, 743–750. [Google Scholar]
- Kimata, Y.; Kohno, K. Endoplasmic reticulum stress-sensing mechanisms in yeast and mammalian cells. Curr. Opin. Cell Biol 2011, 23, 135–142. [Google Scholar]
- Urade, R. The endoplasmic reticulum stress signaling pathways in plants. Biofactors 2009, 35, 326–331. [Google Scholar] [Green Version]
- Iwata, Y.; Koizumi, N. Plant transducers of the endoplasmic reticulum unfolded protein response. Trends Plant Sci 2012, 17, 720–727. [Google Scholar]
- Howell, S.H. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 2013. [Google Scholar] [CrossRef]
- Vitale, A.; Ceriotti, A. Protein quality control mechanisms and protein storage in the endoplasmic reticulum. A conflict of interests? Plant Physiol 2004, 136, 3420–3426. [Google Scholar]
- Tanaka, K.; Sugimoto, T.; Ogawa, M.; Kasai, Z. Isolation and characterization of two types of protein bodies in the rice endosperm. Agric. Biol. Chem 1980, 44, 1633–1639. [Google Scholar]
- Takagi, H.; Hiroi, T.; Yang, L.; Tada, Y.; Yuki, Y.; Takamura, K.; Ishimitsu, R.; Kawauchi, H.; Kiyono, H.; Takaiwa, F. A rice-based edible vaccine expressing multiple T cell epitopes induces oral tolerance for inhibition of Th2-mediated IgE responses. Proc. Natl. Acad. Sci. USA 2005, 102, 17525–17530. [Google Scholar]
- Takaiwa, F.; Takagi, H.; Hirose, S.; Wakasa, Y. Endosperm tissue is good production platform for artificial recombinant proteins in transgenic rice. Plant Biotechnol. J 2007, 5, 84–92. [Google Scholar]
- Oono, Y.; Wakasa, Y.; Hirose, S.; Yang, L.; Sakuta, C.; Takaiwa, F. Analysis of ER stress in developing rice endosperm accumulating beta-amyloid peptide. Plant Biotechnol. J 2010, 8, 691–718. [Google Scholar]
- Wakasa, Y.; Yasuda, H.; Oono, Y.; Kawakatsu, T.; Hirose, S.; Takahashi, H.; Hayashi, S.; Yang, L.; Takaiwa, F. Expression of ER quality control-related genes in response to changes in BiP1 levels in developing rice endosperm. Plant J 2011, 65, 675–689. [Google Scholar]
- Wakasa, Y.; Hayashi, S.; Takaiwa, F. Expression of OsBiP4 and OsBiP5 is highly correlated with the endoplasmic reticulum stress response in rice. Planta 2012, 236, 1519–1527. [Google Scholar]
- Wang, D.; Weaver, N.D.; Kesarwani, M.; Dong, X. Induction of protein secretory pathway is required for systemic acquired resistance. Science 2005, 308, 1036–1040. [Google Scholar]
- Nekrasov, V.; Li, J.; Batoux, M.; Roux, M.; Chu, Z.-H.; Lacombe, S.; Rougon, A.; Bittel, P.; Kiss-Papp, M.; Chinchilla, D.; et al. Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J 2009, 28, 3428–3438. [Google Scholar]
- Saijo, Y.; Tintor, N.; Lu, X.; Rauf, P.; Pajerowska-Mukhtar, K.; Häweker, H.; Dong, X.; Robatzek, S.; Schulze-Lefert, P. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J 2009, 28, 3439–3449. [Google Scholar]
- Wakasa, Y.; Yasuda, H.; Takaiwa, F. Secretory type of recombinant thioredoxin h induces ER stress in endosperm cells of transgenic rice. J. Plant Physiol 2013, 170, 202–210. [Google Scholar]
- Kudo, K.; Ohta, M.; Yang, L.; Wakasa, Y.; Takahashi, S.; Takaiwa, F. ER stress response induced by the production of human IL-7 in rice endosperm cells. Plant Mol. Biol 2013, 81, 461–475. [Google Scholar]
- Watanabe, N.; Lam, E. BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J. Biol. Chem 2008, 283, 3200–3210. [Google Scholar]
- Hayashi, S.; Takahashi, H.; Wakasa, Y.; Kawakatsu, T.; Takaiwa, F. Identification of a cis-element that mediates multiple pathways of the ER stress response in rice. Plant J 2013, 74, 248–257. [Google Scholar]
- Takahashi, H.; Kawakatsu, T.; Wakasa, Y.; Hayashi, S.; Takaiwa, F. A rice transmembrane bZIP transcription factor, OsbZIP39, regulates the endoplasmic reticulum stress response. Plant Cell Physiol 2012, 53, 144–153. [Google Scholar]
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol 2007, 8, 519–529. [Google Scholar]
- Koizumi, N.; Martinez, I.M.; Kimata, Y.; Kohno, K.; Sano, H.; Chrispeels, M.J. Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol 2001, 127, 949–962. [Google Scholar]
- Okushima, Y.; Koizumi, N.; Yamaguchi, Y.; Kimata, Y.; Kohno, K.; Sano, H. Isolation and characterization of a putative transducer of endoplasmic reticulum stress in Oryza sativa. Plant Cell Physiol 2002, 43, 532–539. [Google Scholar]
- Liu, J.-X.; Srivastava, R.; Che, P.; Howell, S.H. An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 2007, 19, 4111–4119. [Google Scholar]
- Mori, K.; Ogawa, N.; Kawahara, T.; Yanagi, H.; Yura, T. mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc. Natl. Acad. Sci. USA 2000, 97, 4660–4665. [Google Scholar]
- Yoshida, H.; Matsui, T.; Yamamoto, A.; Okada, T.; Mori, K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 2001, 107, 881–891. [Google Scholar]
- Deng, Y.; Humbert, S.; Liu, J.-X.; Srivastava, R.; Rothstein, S.J.; Howell, S.H. Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 7247–7252. [Google Scholar]
- Nagashima, Y.; Mishiba, K.-I.; Suzuki, E.; Shimada, Y.; Iwata, Y.; Koizumi, N. Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci. Rep 2011, 1, 29. [Google Scholar]
- Hayashi, S.; Wakasa, Y.; Takahashi, H.; Kawakatsu, T.; Takaiwa, F. Signal transduction by IRE1-mediated splicing of bZIP50 and other stress sensors in the endoplasmic reticulum stress response of rice. Plant J 2012, 69, 946–956. [Google Scholar]
- Hetz, C.; Glimcher, L.H. Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome. Mol. Cell 2009, 35, 551–561. [Google Scholar]
- Wakasa, Y.; Hayashi, S.; Ozawa, K.; Takaiwa, F. Multiple roles of the ER stress sensor IRE1 demonstrated by gene targeting in rice. Sci. Rep 2012, 2, 944. [Google Scholar]
- Haze, K.; Yoshida, H.; Yanagi, H.; Yura, T.; Mori, K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 1999, 10, 3787–3799. [Google Scholar]
- Brown, M.S.; Ye, J.; Rawson, R.B.; Goldstein, J.L. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell 2000, 100, 391–398. [Google Scholar]
- Che, P.; Bussell, J.D.; Zhou, W.; Estavillo, G.M.; Pogson, B.J.; Smith, S.M. Signaling from the endoplasmic reticulum activates brassinosteroid signaling and promotes acclimation to stress in Arabidopsis. Sci. Signal. 2010, 3, ra69. [Google Scholar]
- Liu, J.-X.; Howell, S.H. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 2010, 22, 782–796. [Google Scholar]
- Martínez, I.M.; Chrispeels, M.J. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 2003, 15, 561–576. [Google Scholar]
- Yamamoto, K.; Yoshida, H.; Kokame, K.; Kaufman, R.J.; Mori, K. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J. Biochem 2004, 136, 343–350. [Google Scholar]
- Oh, D.-H.; Kwon, C.-S.; Sano, H.; Chung, W.-I.; Koizumi, N. Conservation between animals and plants of the cis-acting element involved in the unfolded protein response. Biochem. Biophys. Res. Commun 2003, 301, 225–230. [Google Scholar]
- Iwata, Y.; Yoneda, M.; Yanagawa, Y.; Koizumi, N. Characteristics of the nuclear form of the Arabidopsis transcription factor AtbZIP60 during the endoplasmic reticulum stress response. Biosci. Biotechnol. Biochem 2009, 73, 865–869. [Google Scholar]
- Kamauchi, S.; Nakatani, H.; Nakano, C.; Urade, R. Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J 2005, 272, 3461–3476. [Google Scholar]
- Hayashi, S.; Wakasa, Y.; Takaiwa, F. Functional integration between defence and IRE1-mediated ER stress response in rice. Sci. Rep 2012, 2, 670. [Google Scholar]
- Nakagawa, H.; Ohmiya, K.; Hattori, T. A rice bZIP protein, designated OSBZ8, is rapidly induced by abscisic acid. Plant J 1996, 9, 217–227. [Google Scholar]
- Hollien, J.; Weissman, J.S. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 2006, 313, 104–107. [Google Scholar]
- Mishiba, K.-I.; Nagashima, Y.; Suzuki, E.; Hayashi, N.; Ogata, Y.; Shimada, Y.; Koizumi, N. Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc. Natl. Acad. Sci. USA 2013, (in press). [Google Scholar]
- Hong, Z.; Jin, H.; Fitchette, A.-C.; Xia, Y.; Monk, A.M.; Faye, L.; Li, J. Mutations of an alpha1,6 mannosyltransferase inhibit endoplasmic reticulum-associated degradation of defective brassinosteroid receptors in Arabidopsis. Plant Cell 2009, 21, 3792–3802. [Google Scholar]
- Liu, Y.; Burgos, J.S.; Deng, Y.; Srivastava, R.; Howell, S.H.; Bassham, D.C. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 2012, 24, 4635–4651. [Google Scholar]
- Shimono, M.; Sugano, S.; Nakayama, A.; Jiang, C.-J.; Ono, K.; Toki, S.; Takatsuji, H. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 2007, 19, 2064–2076. [Google Scholar]
- Moreno, A.A.; Mukhtar, M.S.; Blanco, F.; Boatwright, J.L.; Moreno, I.; Jordan, M.R.; Chen, Y.; Brandizzi, F.; Dong, X.; Orellana, A.; et al. IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS One 2012, 7, e31944. [Google Scholar]
- Ye, C.; Dickman, M.B.; Whitham, S.A.; Payton, M.; Verchot, J. The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol 2011, 156, 741–755. [Google Scholar]
- Yang, J.; Zhao, X.; Cheng, K.; Du, H.; Ouyang, Y.; Chen, J.; Qiu, S.; Huang, J.; Jiang, Y.; Jiang, L.; et al. A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science 2012, 337, 1336–1340. [Google Scholar]




© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hayashi, S.; Wakasa, Y.; Takaiwa, F. Recent Advances in Understanding the Control of Secretory Proteins by the Unfolded Protein Response in Plants. Int. J. Mol. Sci. 2013, 14, 9396-9407. https://doi.org/10.3390/ijms14059396
Hayashi S, Wakasa Y, Takaiwa F. Recent Advances in Understanding the Control of Secretory Proteins by the Unfolded Protein Response in Plants. International Journal of Molecular Sciences. 2013; 14(5):9396-9407. https://doi.org/10.3390/ijms14059396
Chicago/Turabian StyleHayashi, Shimpei, Yuhya Wakasa, and Fumio Takaiwa. 2013. "Recent Advances in Understanding the Control of Secretory Proteins by the Unfolded Protein Response in Plants" International Journal of Molecular Sciences 14, no. 5: 9396-9407. https://doi.org/10.3390/ijms14059396
APA StyleHayashi, S., Wakasa, Y., & Takaiwa, F. (2013). Recent Advances in Understanding the Control of Secretory Proteins by the Unfolded Protein Response in Plants. International Journal of Molecular Sciences, 14(5), 9396-9407. https://doi.org/10.3390/ijms14059396