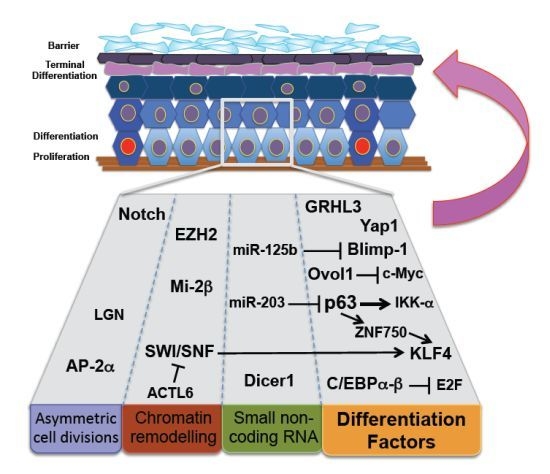
Stem Cells behind the Barrier
Abstract
:1. Introduction
2. Stem Cell Differentiation and Skin Homeostasis
3. Asymmetric Cell Division and Epidermal Barrier Function
4. Terminal Differentiating Factors and Stem Cell Differentiation
5. MicroRNA Regulation of Epidermal Homeostasis
6. Chromatin Remodeling Complexes and Differentiation Programs
7. Differentiation Defects and Epidermal Cancers
8. Conclusions
Acknowledgments
Conflict of Interest
References
- Boehnke, K.; Falkowska-Hansen, B.; Stark, H.J.; Boukamp, P. Stem cells of the human epidermis and their niche: Composition and function in epidermal regeneration and carcinogenesis. Carcinogenesis 2012, 33, 1247–1258. [Google Scholar]
- Moore, K.A.; Lemischka, I.R. Stem cells and their niches. Science 2006, 311, 1880–1885. [Google Scholar]
- Lechler, T.; Fuchs, E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 2005, 437, 275–280. [Google Scholar]
- Klein, A.M.; Doupé, D.P.; Jones, P.H.; Simons, B.D. Kinetics of cell division in epidermal maintenance. Phys. Rev. E 2007, 76, 021910. [Google Scholar]
- Mascre, G.; Dekoninck, S.; Drogat, B.; Youssef, K.K.; Broheé, S.; Sotiropoulou, P.A.; Simons, B.D.; Blanpain, C. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature 2012, 489, 257–262. [Google Scholar]
- Blanpain, C.; Horsley, V.; Fuchs, E. Epithelial stem cells: Turning over new leaves. Cell 2007, 128, 445–458. [Google Scholar]
- Bickenbach, J.R. Identification and behavior of label-retaining cells in oral mucosa and skin. J. Dent. Res 1981, 60, 1611–1620. [Google Scholar]
- Tumbar, T.; Guasch, G.; Greco, V.; Blanpain, C.; Lowry, W.E.; Rendl, M.; Fuchs, E. Defining the epithelial stem cell niche in skin. Science 2004, 303, 359–363. [Google Scholar]
- Jaks, V.; Barker, N.; Kasper, M.; van Es, J.H.; Snippert, H.J.; Clevers, H.; Toftgård, R. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet 2008, 40, 1291–1299. [Google Scholar]
- Morris, R.J.; Liu, Y.; Marles, L.; Yang, Z.; Trempus, C.; Li, S.; Lin, J.S.; Sawicki, J.A.; Cotsarelis, G. Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol 2004, 22, 411–417. [Google Scholar]
- Trempus, C.S.; Morris, R.J.; Bortner, C.D.; Cotsarelis, G.; Faircloth, R.S.; Reece, J.M.; Tennant, R.W. Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. J. Invest. Dermatol 2003, 120, 501–511. [Google Scholar]
- Blanpain, C.; Lowry, W.E.; Geoghegan, A.; Polak, L.; Fuchs, E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 2004, 118, 635–648. [Google Scholar]
- Jones, P.H.; Harper, S.; Watt, F.M. Stem cell patterning and fate in human epidermis. Cell 1995, 80, 83–93. [Google Scholar]
- Li, A.; Simmons, P.J.; Kaur, P. Identification and isolation of candidate human keratinocyte stem cells based on cell surface phenotype. Proc. Natl. Acad. Sci. USA 1998, 95, 3902–3907. [Google Scholar]
- Wan, H.; Stone, M.G.; Simpson, C.; Reynolds, L.E.; Marshall, J.F.; Hart, I.R.; Hodivala-Dilke, K.M.; Eady, R.A. Desmosomal proteins, including desmoglein 3, serve as novel negative markers for epidermal stem cell-containing population of keratinocytes. J. Cell Sci 2003, 116, 4239–4248. [Google Scholar]
- Fortunel, N.O.; Hatzfeld, J.A.; Rosemary, P.A.; Ferraris, C.; Monier, M.N.; Haydont, V.; Longuet, J.; Brethon, B.; Lim, B.; Castiel, I.; et al. Long-term expansion of human functional epidermal precursor cells: Promotion of extensive amplification by low TGF-beta1 concentrations. J. Cell Sci 2003, 116, 4043–4052. [Google Scholar]
- Legg, J.; Jensen, U.B.; Broad, S.; Leigh, I.; Watt, F.M. Role of melanoma chondroitin sulphate proteoglycan in patterning stem cells in human interfollicular epidermis. Development 2003, 130, 6049–6063. [Google Scholar]
- Niemann, C.; Watt, F.M. Designer skin: Lineage commitment in postnatal epidermis. Trends Cell Biol 2002, 12, 185–192. [Google Scholar]
- Watt, F.M.; Jensen, K.B. Epidermal stem cell diversity and quiescence. EMBO Mol. Med 2009, 1, 260–267. [Google Scholar]
- Ito, M.; Yang, Z.; Andl, T.; Cui, C.; Kim, N.; Millar, S.E.; Cotsarelis, G. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature 2007, 447, 316–320. [Google Scholar]
- Castilho, R.M.; Squarize, C.H.; Chodosh, L.A.; Williams, B.O.; Gutkind, J.S. mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell 2009, 5, 279–289. [Google Scholar]
- Li, L.; Clevers, H. Coexistence of quiescent and active adult stem cells in mammals. Science 2010, 327, 542–545. [Google Scholar]
- Horsley, V.; Aliprantis, A.O.; Polak, L.; Glimcher, L.H.; Fuchs, E. NFATc1 balances quiescence and proliferation of skin stem cells. Cell 2008, 132, 299–310. [Google Scholar]
- Jensen, K.B.; Collins, C.A.; Nascimento, E.; Tan, D.W.; Frye, M.; Itami, S.; Watt, F.M. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell 2009, 4, 427–439. [Google Scholar]
- Niessen, M.T.; Iden, S.; Niessen, C.M. The in vivo function of mammalian cell and tissue polarity regulators—How to shape and maintain the epidermal barrier. J. Cell Sci 2012, 125, 3501–3510. [Google Scholar]
- Caddy, J.; Wilanowski, T.; Darido, C.; Dworkin, S.; Ting, S.B.; Zhao, Q.; Rank, G.; Auden, A.; Srivastava, S.; Papenfuss, T.A.; et al. Epidermal wound repair is regulated by the planar cell polarity signaling pathway. Dev. Cell 2010, 19, 138–147. [Google Scholar]
- Dworkin, S.; Jane, S.M.; Darido, C. The planar cell polarity pathway in vertebrate epidermal development, homeostasis and repair. Organogenesis 2011, 7, 202–208. [Google Scholar]
- Poulson, N.D.; Lechler, T. Robust control of mitotic spindle orientation in the developing epidermis. J. Cell Biol 2010, 191, 915–922. [Google Scholar]
- Koster, M.I.; Dai, D.; Marinari, B.; Sano, Y.; Costanzo, A.; Karin, M.; Roop, D.R. p63 induces key target genes required for epidermal morphogenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 3255–3260. [Google Scholar]
- Truong, A.B.; Kretz, M.; Ridky, T.W.; Kimmel, R.; Khavari, P.A. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev 2006, 20, 3185–3197. [Google Scholar]
- Williams, S.E.; Beronja, S.; Pasolli, H.A.; Fuchs, E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature 2011, 470, 353–358. [Google Scholar]
- Koster, M.I.; Kim, S.; Huang, J.; Williams, T.; Roop, D.R. TAp63alpha induces AP-2gamma as an early event in epidermal morphogenesis. Dev. Biol 2006, 289, 253–261. [Google Scholar]
- Wang, X.; Pasolli, H.A.; Williams, T.; Fuchs, E. AP-2 factors act in concert with Notch to orchestrate terminal differentiation in skin epidermis. J. Cell Biol 2008, 183, 37–48. [Google Scholar]
- Demehri, S.; Turkoz, A.; Kopan, R. Epidermal Notch1 loss promotes skin tumorigenesis by impacting the stromal microenvironment. Cancer Cell 2009, 16, 55–66. [Google Scholar]
- Dumortier, A.; Durham, A.D.; Di Piazza, M.; Vauclair, S.; Koch, U.; Ferrand, G.; Ferrero, I.; Demehri, S.; Song, L.L.; Farr, A.G.; et al. Atopic dermatitis-like disease and associated lethal myeloproliferative disorder arise from loss of Notch signaling in the murine skin. PLoS One 2010, 5, e9258. [Google Scholar]
- Mills, A.A.; Zheng, B.; Wang, X.J.; Vogel, H.; Roop, D.R.; Bradley, A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 1999, 398, 708–713. [Google Scholar]
- Patel, S.; Xi, Z.F.; Seo, E.Y.; McGaughey, D.; Segre, J.A. Klf4 and corticosteroids activate an overlapping set of transcriptional targets to accelerate in utero epidermal barrier acquisition. Proc. Natl. Acad. Sci. USA 2006, 103, 18668–18673. [Google Scholar]
- Segre, J.A.; Bauer, C.; Fuchs, E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet 1999, 22, 356–360. [Google Scholar]
- Teng, A.; Nair, M.; Wells, J.; Segre, J.A.; Dai, X. Strain-dependent perinatal lethality of Ovol1-deficient mice and identification of Ovol2 as a downstream target of Ovol1 in skin epidermis. Biochim. Biophys. Acta 2007, 1772, 89–95. [Google Scholar]
- Lopez, R.G.; Garcia-Silva, S.; Moore, S.J.; Bereshchenko, O.; Martinez-Cruz, A.B.; Ermakova, O.; Kurz, E.; Paramio, J.M.; Nerlov, C. C/EBPalpha and beta couple interfollicular keratinocyte proliferation arrest to commitment and terminal differentiation. Nat. Cell Biol 2009, 11, 1181–1190. [Google Scholar]
- Magnusdottir, E.; Kalachikov, S.; Mizukoshi, K.; Savitsky, D.; Ishida-Yamamoto, A.; Panteleyev, A.A.; Calame, K. Epidermal terminal differentiation depends on B lymphocyte-induced maturation protein-1. Proc. Natl. Acad. Sci. USA 2007, 104, 14988–14993. [Google Scholar]
- Zhang, H.; Pasolli, H.A.; Fuchs, E. Yes-associated protein (YAP) transcriptional coactivator functions in balancing growth and differentiation in skin. Proc. Natl. Acad. Sci. USA 2011, 108, 2270–2275. [Google Scholar]
- Yu, Z.; Lin, K.K.; Bhandari, A.; Spencer, J.A.; Xu, X.; Wang, N.; Lu, Z.; Gill, G.N.; Roop, D.R.; Wertz, P.; et al. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev. Biol 2006, 299, 122–136. [Google Scholar]
- Senoo, M.; Pinto, F.; Crum, C.P.; McKeon, F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell 2007, 129, 523–536. [Google Scholar]
- Gareus, R.; Huth, M.; Breiden, B.; Nenci, A.; Rösch, N.; Haase, I.; Bloch, W.; Sandhoff, K.; Pasparakis, M. Normal epidermal differentiation but impaired skin-barrier formation upon keratinocyte-restricted IKK1 ablation. Nat. Cell Biol 2007, 9, 461–469. [Google Scholar]
- Sen, G.L.; Boxer, L.D.; Webster, D.E.; Bussat, R.T.; Qu, K.; Zarnegar, B.J.; Johnston, D.; Siprashvili, Z.; Khavari, P.A. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev. Cell 2012, 22, 669–677. [Google Scholar]
- Li, J.; Zheng, H.; Yu, F.; Yu, T.; Liu, C.; Huang, S.; Wang, T.C.; Ai, W. Deficiency of the Kruppel-like factor KLF4 correlates with increased cell proliferation and enhanced skin tumorigenesis. Carcinogenesis 2012, 33, 1239–1246. [Google Scholar]
- Keyes, W.M.; Pecoraro, M.; Aranda, V.; Vernersson-Lindahl, E.; Li, W.; Vogel, H.; Guo, X.; Garcia, E.L.; Michurina, T.V.; Enikolopov, G.N.; et al. DeltaNp63alpha is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell 2011, 8, 164–176. [Google Scholar]
- Nair, M.; Teng, A.; Bilanchone, V.; Agrawal, A.; Li, B.; Dai, X. Ovol1 regulates the growth arrest of embryonic epidermal progenitor cells and represses c-myc transcription. J. Cell Biol 2006, 173, 253–264. [Google Scholar]
- Nascimento, E.M.; Cox, C.L.; MacArthur, S.; Hussain, S.; Trotter, M.; Blanco, S.; Suraj, M.; Nichols, J.; Kübler, B.; Benitah, S.A.; et al. The opposing transcriptional functions of Sin3a and c-Myc are required to maintain tissue homeostasis. Nat. Cell Biol 2011, 13, 1395–1405. [Google Scholar]
- Nerlov, C. The C/EBP family of transcription factors: A paradigm for interaction between gene expression and proliferation control. Trends Cell Biol 2007, 17, 318–324. [Google Scholar]
- Horsley, V.; O’Carroll, D.; Tooze, R.; Ohinata, Y.; Saitou, M.; Obukhanych, T.; Nussenzweig, M.; Tarakhovsky, A.; Fuchs, E. Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell 2006, 126, 597–609. [Google Scholar]
- Sellheyer, K.; Krahl, D. Blimp-1: A marker of terminal differentiation but not of sebocytic progenitor cells. J. Cutan Pathol 2010, 37, 362–370. [Google Scholar]
- Schlegelmilch, K.; Mohseni, M.; Kirak, O.; Pruszak, J.; Vasioukhin, V.; Avruch, J.; Brummelkamp, T.R. Camargo FD Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell 2011, 144, 782–795. [Google Scholar]
- Lian, I.; Kim, J.; Okazawa, H.; Zhao, J.; Zhao, B.; Yu, J.; Chinnaiyan, A.; Israel, M.A.; Goldstein, L.S.; Abujarour, R.; et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev 2010, 24, 1106–1118. [Google Scholar]
- Yi, R.; O’Carroll, D.; Pasolli, H.A.; Zhang, Z.; Dietrich, F.S.; Tarakhovsky, A.; Fuchs, E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet 2006, 38, 356–362. [Google Scholar]
- Mistry, D.S.; Chen, Y.; Sen, G.L. Progenitor function in self-renewing human epidermis is maintained by the exosome. Cell Stem Cell 2012, 11, 127–135. [Google Scholar]
- Auden, A.; Caddy, J.; Wilanowski, T.; Ting, S.B.; Cunningham, J.M.; Jane, S.M. Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expr. Patterns 2006, 6, 964–970. [Google Scholar]
- Ting, S.B.; Wilanowski, T.; Auden, A.; Hall, M.; Voss, A.K.; Thomas, T.; Parekh, V.; Cunningham, J.M.; Jane, S.M. Inositol- and folate-resistant neural tube defects in mice lacking the epithelial-specific factor Grhl-3. Nat. Med 2003, 9, 1513–1519. [Google Scholar]
- Ting, S.B.; Caddy, J.; Hislop, N.; Wilanowski, T.; Auden, A.; Zhao, L.L.; Ellis, S.; Kaur, P.; Uchida, Y.; Holleran, W.M.; et al. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science 2005, 308, 411–413. [Google Scholar]
- Darido, C.; Jane, S.M. Grhl3 and GEF19 in the front rho. Small GTPases 2010, 1, 104–107. [Google Scholar]
- Hislop, N.R.; Caddy, J.; Ting, S.B.; Auden, A.; Vasudevan, S.; King, S.L.; Lindeman, G.J.; Visvader, J.E.; Cunningham, J.M.; Jane, S.M. Grhl3 and Lmo4 play coordinate roles in epidermal migration. Dev. Biol 2008, 321, 263–272. [Google Scholar]
- Darido, C.; Georgy, S.R.; Wilanowski, T.; Dworkin, S.; Auden, A.; Zhao, Q.; Rank, G.; Srivastava, S.; Finlay, M.J.; Papenfuss, A.T.; et al. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell 2011, 20, 635–648. [Google Scholar]
- Zhang, L.; Stokes, N.; Polak, L.; Fuchs, E. Specific microRNAs are preferentially expressed by skin stem cells to balance self-renewal and early lineage commitment. Cell Stem Cell 2011, 8, 294–308. [Google Scholar]
- Yi, R.; Poy, M.N.; Stoffel, M.; Fuchs, E. A skin microRNA promotes differentiation by repressing “stemness”. Nature 2008, 452, 225–229. [Google Scholar]
- Botchkarev, V.A.; Gdula, M.R.; Mardaryev, A.N.; Sharov, A.A.; Fessing, M.Y. Epigenetic regulation of gene expression in keratinocytes. J. Invest. Dermatol 2012, 132, 2505–2521. [Google Scholar]
- Frye, M.; Fisher, A.G.; Watt, F.M. Epidermal stem cells are defined by global histone modifications that are altered by Myc-induced differentiation. PLoS One 2007, 2, e763. [Google Scholar]
- Ezhkova, E.; Pasolli, H.A.; Parker, J.S.; Stokes, N.; Su, I.H.; Hannon, G.; Tarakhovsky, A.; Fuchs, E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 2009, 136, 1122–1135. [Google Scholar]
- Ezhkova, E.; Lien, W.H.; Stokes, N.; Pasolli, H.A.; Silva, J.M.; Fuchs, E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev 2011, 25, 485–498. [Google Scholar]
- Bao, X.; Tang, J.; Lopez-Pajares, V.; Tao, S.; Qu, K.; Crabtree, G.R.; Khavari, P.A. ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF-dependent induction of KLF4. Cell Stem Cell 2013, 12, 193–203. [Google Scholar]
- Indra, A.K.; Dupé, V.; Bornert, J.M.; Messaddeq, N.; Yaniv, M.; Mark, M.; Chambon, P.; Metzger, D. Temporally controlled targeted somatic mutagenesis in embryonic surface ectoderm and fetal epidermal keratinocytes unveils two distinct developmental functions of BRG1 in limb morphogenesis and skin barrier formation. Development 2005, 132, 4533–4544. [Google Scholar]
- Kashiwagi, M.; Morgan, B.A.; Georgopoulos, K. The chromatin remodeler Mi-2beta is required for establishment of the basal epidermis and normal differentiation of its progeny. Development 2007, 134, 1571–1582. [Google Scholar]
- Darido, C.; Buchert, M.; Pannequin, J.; Bastide, P.; Zalzali, H.; Mantamadiotis, T.; Bourgaux, J.F.; Garambois, V.; Jay, P.; Blache, P.; et al. Defective claudin-7 regulation by Tcf-4 and Sox-9 disrupts the polarity and increases the tumorigenicity of colorectal cancer cells. Cancer Res 2008, 68, 4258–4268. [Google Scholar]
- Descargues, P.; Sil, A.K.; Karin, M. IKKalpha, a critical regulator of epidermal differentiation and a suppressor of skin cancer. EMBO J 2008, 27, 2639–2647. [Google Scholar]
- De Cid, R.; Riveira-Munoz, E.; Zeeuwen, P.L.; Robarge, J.; Liao, W.; Dannhauser, E.N.; Giardina, E.; Stuart, P.E.; Nair, R.; Helms, C.; et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat. Genet 2009, 41, 211–215. [Google Scholar]
- Leech, S.N.; Moss, C. A current and online genodermatosis database. Br. J. Dermatol 2007, 156, 1115–1148. [Google Scholar]
- Nestle, F.O.; di Meglio, P.; Qin, J.Z.; Nickoloff, B.J. Skin immune sentinels in health and disease. Nat. Rev. Immunol 2009, 9, 679–691. [Google Scholar]
- Rogers, H.W.; Weinstock, M.A.; Harris, A.R.; Hinckley, M.R.; Feldman, S.R.; Fleischer, A.B.; Coldiron, B.M. Incidence estimate of nonmelanoma skin cancer in the United States, 2006. Arch. Dermatol 2010, 146, 283–287. [Google Scholar]
- Smith, C.H.; Barker, J.N. Psoriasis and its management. BMJ 2006, 333, 380–384. [Google Scholar]
- Kangsamaksin, T.; Park, H.J.; Trempus, C.S.; Morris, R.J. A perspective on murine keratinocyte stem cells as targets of chemically induced skin cancer. Mol. Carcinog 2007, 46, 579–584. [Google Scholar]
- Jensen, K.B.; Jones, J.; Watt, F.M. A stem cell gene expression profile of human squamous cell carcinomas. Cancer Lett 2008, 272, 23–31. [Google Scholar]
- Morris, R.J.; Fischer, S.M.; Slaga, T.J. Evidence that the centrally and peripherally located cells in the murine epidermal proliferative unit are two distinct cell populations. J. Invest. Dermatol 1985, 84, 277–281. [Google Scholar]
- Morris, R.J.; Fischer, S.M.; Slaga, T.J. Evidence that a slowly cycling subpopulation of adult murine epidermal cells retains carcinogen. Cancer Res 1986, 46, 3061–3066. [Google Scholar]
- Youssef, K.K.; van Keymeulen, A.; Lapouge, G.; Beck, B.; Michaux, C.; Achouri, Y.; Sotiropoulou, P.A.; Blanpain, C. Identification of the cell lineage at the origin of basal cell carcinoma. Nat. Cell Biol 2010, 12, 299–305. [Google Scholar]
- Lapouge, G.; Youssef, K.K.; Vokaer, B.; Achouri, Y.; Michaux, C.; Sotiropoulou, P.A.; Blanpain, C. Identifying the cellular origin of squamous skin tumors. Proc. Natl. Acad. Sci. USA 2011, 108, 7431–7436. [Google Scholar]
- Clayton, E.; Doupé, D.P.; Klein, A.M.; Winton, D.J.; Simons, B.D.; Jones, P.H. A single type of progenitor cell maintains normal epidermis. Nature 2007, 446, 185–189. [Google Scholar]
- Ferraris, C.; Chaloin-Dufau, C.; Dhouailly, D. Transdifferentiation of embryonic and postnatal rabbit corneal epithelial cells. Differentiation 1994, 57, 89–96. [Google Scholar]
- Neumuller, R.A.; Knoblich, J.A. Dividing cellular asymmetry: Asymmetric cell division and its implications for stem cells and cancer. Genes Dev 2009, 23, 2675–2699. [Google Scholar]

© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Cangkrama, M.; Ting, S.B.; Darido, C. Stem Cells behind the Barrier. Int. J. Mol. Sci. 2013, 14, 13670-13686. https://doi.org/10.3390/ijms140713670
Cangkrama M, Ting SB, Darido C. Stem Cells behind the Barrier. International Journal of Molecular Sciences. 2013; 14(7):13670-13686. https://doi.org/10.3390/ijms140713670
Chicago/Turabian StyleCangkrama, Michael, Stephen B. Ting, and Charbel Darido. 2013. "Stem Cells behind the Barrier" International Journal of Molecular Sciences 14, no. 7: 13670-13686. https://doi.org/10.3390/ijms140713670
APA StyleCangkrama, M., Ting, S. B., & Darido, C. (2013). Stem Cells behind the Barrier. International Journal of Molecular Sciences, 14(7), 13670-13686. https://doi.org/10.3390/ijms140713670