Untargeted Metabolomics Reveals Predominant Alterations in Lipid Metabolism Following Light Exposure in Broccoli Sprouts
Abstract
:1. Introduction

2. Results and Discussion



3. Experimental Section
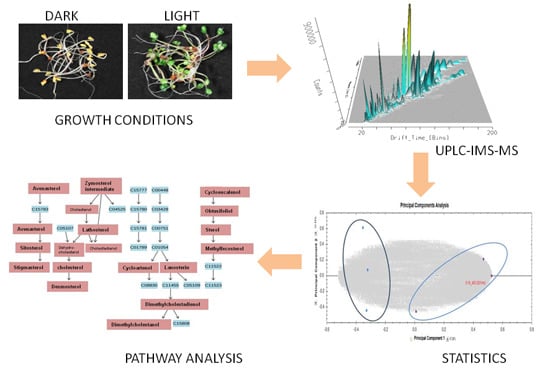
3.1. Growth Conditions
3.2. Sample Preparation
3.3. UPLC Conditions
3.4. MS Conditions
3.5. Data Processing and Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cartea, M.E.; Velasco, P. Glucosinolates in Brassica foods: Bioavailability in food and significance for human health. Phytochem. Rev. 2008, 7, 213–229. [Google Scholar] [CrossRef]
- Traka, M.; Mithen, R. Glucosinolates, isothiocyanates and human health. Phytochem. Rev. 2009, 8, 269–282. [Google Scholar] [CrossRef]
- Verkerk, R.; Schreiner, M.; Krumbein, A.; Ciska, E.; Holst, B.; Rowland, I.; de Schrijver, R.; Hansen, M.; Gerhäuser, C.; Mithen, R. Glucosinolates in Brassica vegetables: The influence of the food supply chain on intake, bioavailability and human health. Mol. Nutr. Food Res. 2009, 53, S219–S219. [Google Scholar] [CrossRef] [PubMed]
- Armah, C.N.; Traka, M.H.; Dainty, J.R.; Defernez, M.; Janssens, A.; Leung, W.; Doleman, J.F.; Potter, J.F.; Mithen, R.F. A diet rich in high-glucoraphanin broccoli interacts with genotype to reduce discordance in plasma metabolite profiles by modulating mitochondrial function. Am. J. Clin. Nutr. 2013, 98, 712–722. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Balibrea, S.; Moreno, D.A.; García-Viguera, C. Influence of light on health-promoting phytochemicals of broccoli sprouts. J. Sci. Food Agric. 2008, 88, 904–910. [Google Scholar] [CrossRef]
- Astarita, G.; Langridge, J. An emerging role for metabolomics in nutrition science. J. Nutrigenet. Nutrigenomics 2013, 6, 181–200. [Google Scholar] [CrossRef] [PubMed]
- Maldini, M.; Baima, S.; Morelli, G.; Scaccini, C.; Natella, F. A liquid chromatography-mass spectrometry approach to study “glucosinoloma” in broccoli sprouts. J. Mass Spectrom. 2012, 47, 1198–1206. [Google Scholar] [CrossRef] [PubMed]
- Aires, A.; Rosa, E.; Carvalho, R. Effect of nitrogen and sulfur fertilization on glucosinolates in the leaves and roots of broccoli sprouts (Brassica oleracea var. italica). J. Sci. Food Agric. 2006, 86, 1512–1516. [Google Scholar] [CrossRef]
- Velasco, P.; Francisco, M.; Moreno, D.A.; Ferreres, F.; García-Viguera, C.; Cartea, M.E. Phytochemical fingerprinting of vegetable Brassica oleracea and Brassica napus by simultaneous identification of glucosinolates and phenolics. Phytochem. Anal. 2011, 22, 144–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, W.T.; Kim, J.K.; Park, S.; Lee, S.-W.; Li, X.; Kim, Y.B.; Uddin, M.R.; Park, N.I.; Kim, S.-J.; Park, S.U. Metabolic profiling of glucosinolates, anthocyanins, carotenoids, and other secondary metabolites in kohlrabi (Brassica oleracea var. gongylodes). J. Agric. Food Chem. 2012, 60, 8111–8116. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Yuan, G.; Wang, Q. Effect of sucrose and mannitol on the accumulation of health-promoting compounds and the activity of metabolic enzymes in broccoli sprouts. Sci. Hortic. 2011, 128, 159–165. [Google Scholar] [CrossRef]
- Guzman, I.; Yousef, G.G.; Brown, A.F. Simultaneous extraction and quantitation of carotenoids, chlorophylls, and tocopherols in brassica vegetables. J. Agric. Food Chem. 2012, 60, 7238–7244. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xiao, Z.; Lin, L.-Z.; Lester, G.E.; Wang, Q.; Harnly, J.M.; Chen, P. Profiling polyphenols in five Brassica species microgreens by UHPLC-PDA-ESI/HRMSn. J. Agric. Food Chem. 2013, 61, 10960–10970. [Google Scholar] [CrossRef] [PubMed]
- Ahmadiani, N.; Robbins, R.J.; Collins, T.M.; Giusti, M.M. Anthocyanins contents, profiles, and color characteristics of red cabbage extracts from different cultivars and maturity stages. J. Agric. Food Chem. 2014, 62, 7524–7531. [Google Scholar] [CrossRef] [PubMed]
- Quanbeck, S.M.; Brachova, L.; Campbell, A.A.; Guan, X.; Perera, A.; He, K.; Rhee, S.Y.; Bais, P.; Dickerson, J.A.; Dixon, P. Metabolomics as a hypothesis-generating functional genomics tool for the annotation of Arabidopsis thaliana genes of “unknown function”. Front. Plant Sci. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics-the link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Martinis, J.; Kessler, F.; Glauser, G. A novel method for prenylquinone profiling in plant tissues by ultra-high pressure liquid chromatography-mass spectrometry. Plant Methods 2011, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Eugeni Piller, L.; Besagni, C.; Ksas, B.; Rumeau, D.; Brehelin, C.; Glauser, G.; Kessler, F.; Havaux, M. Chloroplast lipid droplet type II NAD(P)H quinone oxidoreductase is essential for prenylquinone metabolism and vitamin K1 accumulation. Proc. Natl. Acad. Sci. USA 2011, 108, 14354–14359. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; Williams, J.P.; Menikarachchi, L.; Thompson, J.W.; Tyldesley-Worster, R.; Halldorsson, S.; Rolfsson, O.; Moseley, A.; Grant, D.; Langridge, J.; et al. Ion mobility derived collision cross sections to support metabolomics applications. Anal. Chem. 2014, 86, 3985–3993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzales, G.B.; Raes, K.; Coelus, S.; Struijs, K.; Smagghe, G.; van Camp, J. Ultra (high)-pressure liquid chromatography–electrospray ionization-time-of-flight-ion mobility-high definition mass spectrometry for the rapid identification and structural characterization of flavonoid glycosides from cauliflower waste. J. Chromatogr. A 2014, 1323, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Wang, P.; Meng, X.; Sun, H.; Zhang, A.; Wang, W.; Dong, H.; Wang, X. Ultra-performance Liquid Chromatography—High-definition Mass Spectrometry analysis of constituents in the root of Radix Stemonae and those absorbed in blood after oral administration of the extract of the crude drug. Phytochem. Anal. 2012, 23, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Ni, B.; Zhang, A.; Wang, M.; Dong, H.; Wang, X. Metabolomics study on Fuzi and its processed products using ultra-performance liquid-chromatography/electrospray-ionization synapt high-definition mass spectrometry coupled with pattern recognition analysis. Analyst 2012, 137, 170–185. [Google Scholar] [CrossRef] [PubMed]
- Pacini, T.; Fu, W.; Gudmundsson, S.; Chiaravalle, A.E.; Brynjolfson, S.; Palsson, B.O.; Astarita, G.; Paglia, G. Multidimensional analytical approach based on UHPLC-UV-Ion Mobility-MS for the screening of natural pigments. Anal. Chem. 2015, 87, 2593–2599. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; Angel, P.; Williams, J.P.; Richardson, K.; Olivos, H.J.; Thompson, J.W.; Menikarachchi, L.; Lai, S.; Walsh, C.; Moseley, A.; et al. Ion mobility-derived collision cross section as an additional measure for lipid fingerprinting and identification. Anal. Chem. 2015, 87, 1137–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Baker, A.; Chen, P. Profiling the indole alkaloids in yohimbe bark with ultra-performance liquid chromatography coupled with ion mobility quadrupole time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2011, 25, 2591–2602. [Google Scholar] [PubMed]
- Stopka, S.A.; Shrestha, B.; Maréchal, É.; Falconet, D.; Vertes, A. Metabolic transformation of microalgae due to light acclimation and genetic modifications followed by laser ablation electrospray ionization mass spectrometry with ion mobility separation. Analyst 2014, 139, 5946–5954. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Magnúsdóttir, M.; Brynjólfson, S.; Palsson, B.Ø.; Paglia, G. UPLC-UV-MSE analysis for quantification and identification of major carotenoid and chlorophyll species in algae. Anal. Bioanal. Chem. 2012, 404, 3145–3154. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Mandal, R.; Sinelnikov, I.V.; Broadhurst, D.; Wishart, D.S. MetaboAnalyst 2.0—A comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012, 40, W127–W133. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Xu, Y.; Shang, D.; Yang, H.; Liu, W.; Han, J.; Sun, Z.; Yao, Q.; Zhang, C.; Ma, J.; et al. MPINet: Metabolite pathway identification via coupling of global metabolite network structure and metabolomic profile. Biomed. Res. Int. 2014, 2014, 325697. [Google Scholar] [CrossRef] [PubMed]
- Cavill, R.; Kamburov, A.; Ellis, J.K.; Athersuch, T.J.; Blagrove, M.S.; Herwig, R.; Ebbels, T.M.; Keun, H.C. Consensus-phenotype integration of transcriptomic and metabolomic data implies a role for metabolism in the chemosensitivity of tumour cells. PLoS Comput. Biol. 2011, 7, e1001113. [Google Scholar] [CrossRef] [PubMed]
- Chory, J.; Chatterjee, M.; Cook, R.; Elich, T.; Fankhauser, C.; Li, J.; Nagpal, P.; Neff, M.; Pepper, A.; Poole, D. From seed germination to flowering, light controls plant development via the pigment phytochrome. Proc. Natl. Acad. Sci. USA 1996, 93, 12066–12071. [Google Scholar] [CrossRef] [PubMed]
- Clouse, S.D.; Sasse, J.M. Brassinosteroids: Essential regulators of plant growth and development. Annu. Rev. Plant Biol. 1998, 49, 427–451. [Google Scholar] [CrossRef] [PubMed]
- Phillip, D.; Ruban, A.V.; Horton, P.; Asato, A.; Young, A.J. Quenching of chlorophyll fluorescence in the major light-harvesting complex of photosystem II: A systematic study of the effect of carotenoid structure. Proc. Natl. Acad. Sci. USA 1996, 93, 1492–1497. [Google Scholar] [CrossRef] [PubMed]
- Vershinin, A. Biological functions of carotenoids–diversity and evolution. Biofactors 1999, 10, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Klyachko-Gurvich, G.L.; Tsoglin, L.N.; Doucha, J.; Kopetskii, J.; Semenenko, V.E. Desaturation of fatty acids as an adaptive response to shifts in light intensity 1. Physiol. Plant. 1999, 107, 240–249. [Google Scholar] [CrossRef]
- Gombos, Z.; Wada, H.; Hideg, E.; Murata, N. The unsaturation of membrane lipids stabilizes photosynthesis against heat stress. Plant Physiol. 1994, 104, 563–567. [Google Scholar] [PubMed]
- Calder, P.C.; Yaqoob, P. Understanding omega-3 polyunsaturated fatty acids. Postgrad. Med. 2009, 121, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Simopoulos, A.P. Essential fatty acids in health and chronic disease. Am. J. Clin. Nutr. 1999, 70, 560s–569s. [Google Scholar] [PubMed]
- Sies, H.; Stahl, W. Non-nutritive bioactive food constituents of plants: Lycopene, lutein and zeaxanthin. Int. J. Vitam. Nutr. Res. 2003, 73, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Stahl, W.; Sies, H. Bioactivity and protective effects of natural carotenoids. Biochim. Biophys. Acta 2005, 1740, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.J.; MacDougall, D.E.; Ntanios, F.; Vanstone, C.A. Dietary phytosterols as cholesterol-lowering agents in humans. Can. J. Physiol. Pharmacol. 1997, 75, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Awad, A.B.; Fink, C.S. Phytosterols as anticancer dietary components: Evidence and mechanism of action. J. Nutr. 2000, 130, 2127–2130. [Google Scholar] [PubMed]
- Glueck, C.J.; Speirs, J.; Tracy, T.; Streicher, P.; Illig, E.; Vandegrift, J. Relationships of serum plant sterols (phytosterols) and cholesterol in 595 hypercholesterolemic subjects, and familial aggregation of phytosterols, cholesterol, and premature coronary heart disease in hyperphytosterolemic probands and their first-degree relatives. Metabolism 1991, 40, 842–848. [Google Scholar] [PubMed]
- Hollman, P.C.H.; Katan, M. Dietary flavonoids: Intake, health effects and bioavailability. Food Chem. Toxicol. 1999, 37, 937–942. [Google Scholar] [CrossRef]
- Plumb, G.W.; Price, K.R.; Modes, M.J.; Williamson, G. Antioxidant properties of the major polyphenolic compounds in broccoli. Free Radic. Res. 1997, 27, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Gorelik, S.; Lapidot, T.; Shaham, I.; Granit, R.; Ligumsky, M.; Kohen, R.; Kanner, J. Lipid peroxidation and coupled vitamin oxidation in simulated and human gastric fluid inhibited by dietary polyphenols: Health implications. J. Agric. Food Chem. 2005, 53, 3397–3402. [Google Scholar] [CrossRef] [PubMed]
- Cartea, M.E.; Francisco, M.; Soengas, P.; Velasco, P. Phenolic compounds in Brassica vegetables. Molecules 2011, 16, 251–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jahangir, M.; Abdel-Farid, I.B.; Choi, Y.H.; Verpoorte, R. Metal ion-inducing metabolite accumulation in Brassica rapa. J. Plant Physiol. 2008, 165, 1429–1437. [Google Scholar] [CrossRef] [PubMed]
- Podsędek, A. Natural antioxidants and antioxidant capacity of Brassica vegetables: A review. LWT-Food Sci. Technol. 2007, 40, 1–11. [Google Scholar] [CrossRef]
- Fahey, J.W.; Zhang, Y.; Talalay, P. Broccoli sprouts: An exceptionally rich source of inducers of enzymes that protect against chemical carcinogens. Proc. Natl. Acad. Sci. USA 1997, 94, 10367–10372. [Google Scholar] [CrossRef] [PubMed]
- Jahangir, M.; Kim, H.K.; Choi, Y.H.; Verpoorte, R. Health-affecting compounds in Brassicaceae. Compr. Rev. Food Sci. Food Saf. 2009, 8, 31–43. [Google Scholar] [CrossRef]
- Zhang, Y. The molecular basis that unifies the metabolism, cellular uptake and chemopreventive activities of dietary isothiocyanates. Carcinogenesis 2012, 33, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Balibrea, S.; Moreno, D.A.; García-Viguera, C. Glucosinolates in broccoli sprouts (Brassica oleracea var. italica) as conditioned by sulphate supply during germination. J. Food Sci. 2010, 75, C673–C677. [Google Scholar] [CrossRef] [PubMed]
- Moreno, D.A.; Carvajal, M.; López-Berenguer, C.; García-Viguera, C. Chemical and biological characterisation of nutraceutical compounds of broccoli. J. Pharm. Biomed. Anal. 2006, 41, 1508–1522. [Google Scholar] [CrossRef] [PubMed]
- Ciska, E.; Martyniak-Przybyszewska, B.; Kozlowska, H. Content of glucosinolates in cruciferous vegetables grown at the same site for two years under different climatic conditions. J. Agric. Food Chem. 2000, 48, 2862–2867. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Balibrea, S.; Moreno, D.A.; García-Viguera, C. Genotypic effects on the phytochemical quality of seeds and sprouts from commercial broccoli cultivars. Food Chem. 2011, 125, 348–354. [Google Scholar] [CrossRef]
- Vallejo, F.; Tomás-Barberán, F.; García-Viguera, C. Glucosinolates and vitamin C content in edible parts of broccoli florets after domestic cooking. Eur. Food Res. Technol. 2002, 215, 310–316. [Google Scholar]
- Goodspeed, D.; Liu, J.D.; Chehab, E.W.; Sheng, Z.; Francisco, M.; Kliebenstein, D.J.; Braam, J. Postharvest circadian entrainment enhances crop pest resistance and phytochemical cycling. Curr. Biol. 2013, 23, 1235–1241. [Google Scholar] [CrossRef] [PubMed]
- Talalay, P.; Fahey, J.W. Phytochemicals from cruciferous plants protect against cancer by modulating carcinogen metabolism. J. Nutr. 2001, 131, 3027S–3033S. [Google Scholar] [PubMed]
- McConn, M. The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell Online 1996, 8, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Berdyshev, E.V. Mass spectrometry of fatty aldehydes. Biochim. Biophys. Acta 2011, 1811, 680–693. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Subramaniam, S.; Murphy, R.C.; Nishijima, M.; Raetz, C.R.; Shimizu, T.; Spener, F.; van Meer, G.; Wakelam, M.J.; Dennis, E.A. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009, 50, S9–S14. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maldini, M.; Natella, F.; Baima, S.; Morelli, G.; Scaccini, C.; Langridge, J.; Astarita, G. Untargeted Metabolomics Reveals Predominant Alterations in Lipid Metabolism Following Light Exposure in Broccoli Sprouts. Int. J. Mol. Sci. 2015, 16, 13678-13691. https://doi.org/10.3390/ijms160613678
Maldini M, Natella F, Baima S, Morelli G, Scaccini C, Langridge J, Astarita G. Untargeted Metabolomics Reveals Predominant Alterations in Lipid Metabolism Following Light Exposure in Broccoli Sprouts. International Journal of Molecular Sciences. 2015; 16(6):13678-13691. https://doi.org/10.3390/ijms160613678
Chicago/Turabian StyleMaldini, Mariateresa, Fausta Natella, Simona Baima, Giorgio Morelli, Cristina Scaccini, James Langridge, and Giuseppe Astarita. 2015. "Untargeted Metabolomics Reveals Predominant Alterations in Lipid Metabolism Following Light Exposure in Broccoli Sprouts" International Journal of Molecular Sciences 16, no. 6: 13678-13691. https://doi.org/10.3390/ijms160613678
APA StyleMaldini, M., Natella, F., Baima, S., Morelli, G., Scaccini, C., Langridge, J., & Astarita, G. (2015). Untargeted Metabolomics Reveals Predominant Alterations in Lipid Metabolism Following Light Exposure in Broccoli Sprouts. International Journal of Molecular Sciences, 16(6), 13678-13691. https://doi.org/10.3390/ijms160613678