Global Transcriptomic Analysis of Targeted Silencing of Two Paralogous ACC Oxidase Genes in Banana
Abstract
:1. Introduction
2. Results
2.1. Chromosomal Distribution of the 1-Aminocyclopropane-1-carboxylic acid (ACC) Oxidase Genes on the Banana Genome
2.2. Generation of Two RNA Interference Transgenic Banana Plants by Expressing siRNA for Mh-ACO1 and Mh-ACO2
2.3. Isolation of Total RNA from Mh-ACO1 and Mh-ACO2 RNAi Plants for RNA-Sequence Analysis
2.4. Assembly of Reads and Transcriptome Characterization of Mh-ACO1 and Mh-ACO2 RNAi Plants by High-Throughput RNA Sequencing
2.5. Transcriptome Characterization of Mh-ACO1 and Mh-ACO2 RNAi Plants by High-Throughput RNA Sequencing
2.5.1. Gene Oncology (GO) Assignments
2.5.2. Eukaryotic Orthologous Groups (KOG) Annotation
2.5.3. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Mapping
2.5.4. Gene Expression Difference Analysis
2.5.5. GO Enrichment Analysis of Differentially Expressed Genes (DEGs)
2.5.6. KEGG Pathway Enrichment Analysis
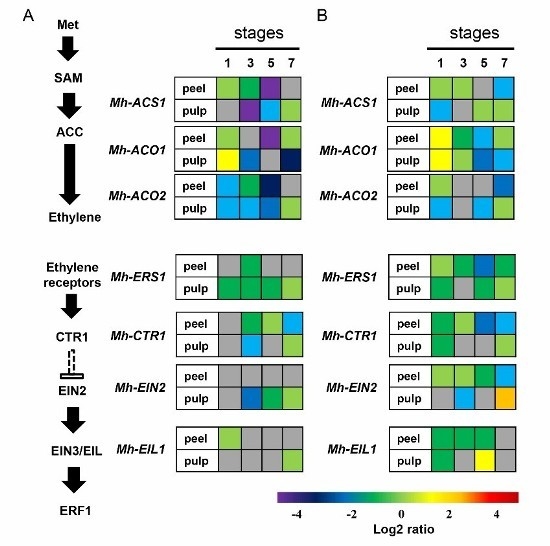
2.6. Expression of the Genes Involved in Ethylene Biosynthesis and Signal Transduction Pathway in Mh-ACO1 and Mh-ACO2 RNAi Transgenic Banana Fruits
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Plasmid Construction and Banana Transformation
4.3. Southern Blot Analysis of Transgenic Banana Plants
4.4. Total RNA Extraction and Whole-Transcriptome Deep Sequencing
4.5. Quantitative Real-Time PCR
4.6. R Programming for Heatmap Illustration
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Beaudry, R.M.; Paz, N.; Black, C.C.; Kays, S.J. Banana ripening: implications of changes in internal ethylene and CO2 concentrations, pulp fructose 2,6-bisphosphate concentration, and activity of some glycolytic enzymes. Plant Physiol. 1987, 85, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Beaudry, R.M.; Severson, R.F.; Black, C.C.; Kays, S.J. Banana ripening: implications of changes in glycolytic intermediate concentrations, glycolytic and gluconeogenic carbon flux, and fructose 2,6-bisphosphate concentration. Plant Physiol. 1989, 91, 1436–1444. [Google Scholar] [CrossRef] [PubMed]
- Bleecker, A.; Kende, H. Ethylene: A gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 2000, 16, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Alexander, L.; Grierson, D. Ethylene biosynthesis and action in tomato: A model for climateric fruit ripening. J. Exp. Bot. 2002, 53, 2039–2055. [Google Scholar] [CrossRef] [PubMed]
- Zarembinski, T.I.; Theologis, A. Ethylene biosynthesis and action: A case of conservation. Plant Mol. Biol. 1994, 26, 1579–1597. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S.R.; Roy, S.; Saha, P.P.; Singh, S.K.; Sengupta, D.N. Characterization of differential ripening pattern in association with ethylene biosynthesis in the fruits of five naturally occurring banana cultivars and detection of a GCC-box-specific DNA-binding protein. Plant Cell Rep. 2008, 27, 1235–1249. [Google Scholar] [CrossRef] [PubMed]
- Parrish, D.W.; Sharma, S.; Kumar, S. Characterization of ethylene biosynthesis associated with ripening in banana fruit. Plant Physiol. 1999, 121, 1257–1265. [Google Scholar]
- Yang, S.F.; Hoffman, N.E. Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 2003, 35, 155–189. [Google Scholar] [CrossRef]
- Rottmann, W.H.; Peter, G.F.; Oeller, P.W.; Keller, J.A.; Shen, N.F.; Nagy, B.P.; Taylor, L.P.; Campbell, A.D.; Theologis, A. 1-Aminocyclopropane-1-carboxylate synthase in tomato is encoded by a multigene family whose transcription is induced during fruit and floral senescence. J. Mol. Biol. 1991, 222, 937–961. [Google Scholar] [CrossRef]
- Sato, T.; Theologis, A. Cloning the mRNA encoding 1-aminocyclopropane-1-carboxylate synthase, the key enzyme for ethylene biosynthesis in plants. Proc. Natl. Acad. Sci. USA 1989, 86, 6621–6625. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.G.; Kim, W.T.; Yip, W.K.; Thompson, G.A.; Li, L.; Bennett, A.B.; Yang, S.F. Cloning of a cDNA encoding 1-aminocyclopropane-1-carboxylate synthase and expression of its mRNA in ripening apple fruit. Planta 1991, 185, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Neupane, K.R.; Mukatira, U.T.; Kato, C.; Stiles, J.I. Cloning and characterization of fruit-expressed ACC synthase and ACC oxidase from papaya (Carica. papaya L.). Acta Hortic. 1998, 461, 329–338. [Google Scholar] [CrossRef]
- Ikoma, Y.; Yano, M.; Ogawa, K. Cloning and expression of genes encoding ACC synthase in kiwifruit. Acta Hortic. 1995, 398, 179–186. [Google Scholar] [CrossRef]
- Wang, A.Q.; Yang, L.T.; Wang, Z.Z.; Wei, Y.T.; He, L.F.; Li, Y.R. Cloning of three members of ACC synthase gene family in sugarcane. Sugar Tech 2013, 8, 257–263. [Google Scholar] [CrossRef]
- Zhang, C.; Shen, Z.; Zhang, Y.; Jian, H.; Ma, R.; Korir, N.K.; Yu, M. Cloning and expression of genes related to the sucrose-metabolizing enzymes and carbohydrate changes in peach. Acta Physiol. Plant. 2013, 35, 589–602. [Google Scholar] [CrossRef]
- Huang, P.L.; Do, Y.Y.; Huang, F.C.; Thay, T.S.; Chang, T.W. Characterization and expression analysis of a banana gene encoding 1-aminocyclopropane-1-carboxylate oxidase. Biochem. Mol. Biol. Int. 1997, 41, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Van Der Straeten, D.; Van Wiemeersch, L.; Goodman, H.M.; Van Montagu, M. Cloning and sequence of two different cDNAs encoding 1-aminocyclopropane-1-carboxylate synthase in tomato. Proc. Natl. Acad. Sci. USA 1990, 87, 4859–4863. [Google Scholar] [CrossRef] [PubMed]
- Pariasca, J.A.T.; Sunaga, A.; Miyazaki, T.; Hisaka, H.; Sonoda, M.; Nakagawa, H.; Sato, T. Cloning of cDNAs encoding senescence-associated genes, ACC synthase and ACC oxidase from stored snow pea pods (Pisum. sativum L. var. saccharatum) and their expression during pod storage. Postharvest Biol. Technol. 2001, 22, 239–247. [Google Scholar] [CrossRef]
- Alonso, J.M.; Stepanova, A.N.; Solano, R.; Wisman, E.; Ferrari, S.; Ausubel, F.M.; Ecker, J.R. Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 2992–2997. [Google Scholar] [CrossRef] [PubMed]
- Hua, J.; Chang, C.; Sun, Q.; Meyerowitz, E.M. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science 1995, 269, 1712–1714. [Google Scholar] [CrossRef] [PubMed]
- Hua, J.; Meyerowitz, E.M. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 1998, 94, 261–271. [Google Scholar] [CrossRef]
- Kieber, J.J.; Rothenberg, M.; Roman, G.; Feldmann, K.A.; Ecker, J.R. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 1993, 72, 427–441. [Google Scholar] [CrossRef]
- Alonso, J.M.; Ecker, J.R. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 1999, 284, 2148–2152. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.L.; Larsen, P.B.; Wang, X.X.; Chang, C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc. Natl. Acad. Sci. USA 1998, 95, 5401–5406. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ecker, J.R. Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 2003, 115, 667–677. [Google Scholar] [CrossRef]
- Inaba, A.; Liu, X.; Yokotan, N.; Yamane, M.; Lu, W.J.; Nakano, R.; Kubo, Y. Differential feedback regulation of ethylene biosynthesis in pulp and peel tissues of banana fruit. J. Exp. Bot. 2007, 58, 1047–1057. [Google Scholar] [CrossRef] [PubMed]
- Kende, H. Ethylene Biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993, 44, 283–307. [Google Scholar] [CrossRef]
- Do, Y.Y.; Thay, T.S.; Chang, T.W.; Huang, P.L. Molecular cloning and characterization of a novel 1-aminocyclopropane-1-carboxylate oxidase gene involved in ripening of banana fruits. J. Agric. Food Chem. 2005, 53, 8239–8247. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.W.; Gudimella, R.; Harikrishna, J.A.; Sin, L.W.; Khalid, N.; Keulemans, J. A draft Musa balbisiana genome sequence for molecular genetics in polyploid, inter- and intra-specific Musa hybrid. BMC Genom. 2013, 14, 683. [Google Scholar] [CrossRef] [PubMed]
- D’Hont, A.; Denoeud, F.; Aury, J.M.; Baurens, F.C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jefferson, R.; Kavanagh, T.; Bevan, M. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [PubMed]
- Banana Genome Hub. Available online: http://banana-genome-hub.southgreen.fr/blast (accessed on 15 June 2016).
- NCBI Gene Expression Omnibus. Available online: http://www.ncbi.nlm.nih.gov/geo/ (accessed on 29 June 2016).
- Blastx2GO. Available online: http://www.blast2go.org/ (accessed on 22 May 2015).
- KEGG. Available online: http://www.genome.jp/kegg/ (accessed on 22 May 2015).
- Swiss-Prot Database. Available online: http://www.ebi.ac.uk/uniprot/ (accessed on 22 May 2015).
- PFAM. Available online: http://pfam.sanger.ac.uk/ (accessed on 22 May 2015).
- GO terms. Available online: http://www.geneontology.org/ (accessed on 22 May 2015).
- KOG. Available online: http://www.ncbi.nlm.nih.gov/COG/ (accessed on 22 May 2015).
- Xiong, A.S.; Yao, Q.H.; Peng, R.H.; Li, X.; Han, P.L.; Fan, H.Q. Different effects on ACC oxidase gene silencing triggered by RNA interference in transgenic tomato. Plant Cell Rep. 2005, 23, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, A.; Zhu, S.; Zhang, L. Expression of ACO1, ERS1 and ERF1 genes in harvested bananas in relation to heat-induced defense against Colletotrichum. musae. J. Plant Physiol. 2011, 168, 1634–1640. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.L.; Do, Y.Y.; Huang, P.L. Expression profiles of a MhCTR1 gene in relation to banana fruit ripening. Plant Physiol. Biochem. 2012, 56, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Dilley, D.R.; Wang, Z.; Kadirjan-Kalbach, D.K.; Ververidis, F.; Beaudry, R.; Padmanabhan, K. 1-Aminocyclopropane-1-carboxylic acid oxidase reaction mechanism and putative post-translational activities of the ACCO protein. AoB Plants 2013, 5, plt031. [Google Scholar] [CrossRef] [PubMed]
- Cordenunsi, B.R.; Lajolo, F.M. Starch breakdown during banana ripening: Sucrose synthase and sucrose phosphate synthase. J. Agric. Food Chem. 1995, 43, 347–351. [Google Scholar] [CrossRef]
- Agrawal, N.; Dasaradhi, P.V.N.; Mohmmed, A.; Malhotra, P.; Bhatnagar, R.K.; Mukherjee, S.K. RNA interference: Biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 2003, 67, 657–685. [Google Scholar] [CrossRef] [PubMed]
- Commonwealth Scientific and Industrial Research Organization (CSIRO) Division of Food Research. Banana Ripening Guide/Division of Food Research; Commonwealth Scientific and Industrial Research Organization: Sydney, Australia, 1972; Volume 8, pp. 1–12. [Google Scholar]
- Hellens, R.P.; Edwards, E.A.; Leyland, N.R.; Bean, S.; Mullineaux, P.M. pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Blol. 2000, 42, 819–832. [Google Scholar] [CrossRef]
- Hsu, S.T.; Liao, Y.W.; Huang, P.L. Establishment of Agrobacterium-mediated transformation system in a cultivated banana (Musa ‘Pei Chiao’, AAA Group). J. Taiwan Soc. Hort. Sci. 2008, 54, 173–181. [Google Scholar]
- Chan, H.T.; Chia, M.Y.; Pang, V.F.; Jeng, C.R.; Do, Y.Y.; Huang, P.L. Oral immunogenicity of porcine reproductive and respiratory syndrome virus antigen expressed in transgenic banana. Plant Biotechnol. J. 2013, 11, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Schenk, R.U.; Hildebrandt, A.C. Medium and techniques for induction and growth of monocotyledonous and dicotyledonous cell cultures. Can. J. Bot. 1972, 50, 199–204. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreparation: Version II. Plant. Mol. Blol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Maretty, L.; Sibbesen, J.A.; Krogh, A. Bayesian transcriptome assembly. Genome Biol. 2014, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and 2−∆∆Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 21 June 2016).










© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, Y.; Kuan, C.; Chiu, C.-H.; Chen, X.-J.; Do, Y.-Y.; Huang, P.-L. Global Transcriptomic Analysis of Targeted Silencing of Two Paralogous ACC Oxidase Genes in Banana. Int. J. Mol. Sci. 2016, 17, 1632. https://doi.org/10.3390/ijms17101632
Xia Y, Kuan C, Chiu C-H, Chen X-J, Do Y-Y, Huang P-L. Global Transcriptomic Analysis of Targeted Silencing of Two Paralogous ACC Oxidase Genes in Banana. International Journal of Molecular Sciences. 2016; 17(10):1632. https://doi.org/10.3390/ijms17101632
Chicago/Turabian StyleXia, Yan, Chi Kuan, Chien-Hsiang Chiu, Xiao-Jing Chen, Yi-Yin Do, and Pung-Ling Huang. 2016. "Global Transcriptomic Analysis of Targeted Silencing of Two Paralogous ACC Oxidase Genes in Banana" International Journal of Molecular Sciences 17, no. 10: 1632. https://doi.org/10.3390/ijms17101632
APA StyleXia, Y., Kuan, C., Chiu, C. -H., Chen, X. -J., Do, Y. -Y., & Huang, P. -L. (2016). Global Transcriptomic Analysis of Targeted Silencing of Two Paralogous ACC Oxidase Genes in Banana. International Journal of Molecular Sciences, 17(10), 1632. https://doi.org/10.3390/ijms17101632