Dimerization of the Vacuolar Receptors AtRMR1 and -2 from Arabidopsis thaliana Contributes to Their Localization in the trans-Golgi Network
Abstract
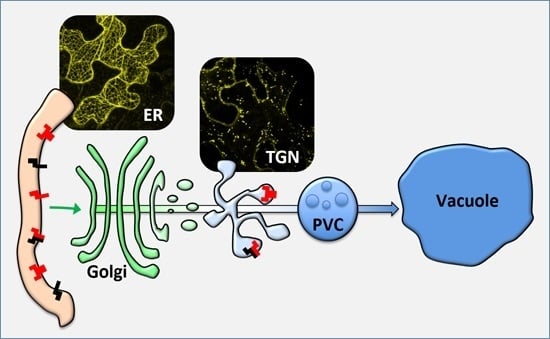
:1. Introduction
2. Results
2.1. AtRMR1 and AtRMR2 Have Different Subcellular Localizations in N. benthamiana Leaves
2.2. The Protease-Associated (PA), RING-H2 and Ser-Rich Domains Are Not Involved in the Subcellular Localization of Either AtRMR1 or AtRMR2
2.3. The Transmembrane Domain Is Not Involved in AtRMRs Localization
2.4. A Short Sequence Linker of AtRMR2 Is Involved in Protein Trafficking to the trans-Golgi Network (TGN)
2.5. AtRMR1 Changes Localization When Co-Expressed with AtRMR2
2.6. Homo- and Hetero-Dimerization of AtRMRs
2.7. AtRMR2 Homodimers Do Not Interact with AtRMR1 to Form Trimers or Tetramers
3. Discussion
3.1. AtRMRs Localize in Different Compartments of the Secretory Pathway
3.2. The Main Protein Domains of AtRMRs, Including the Transmembrane Domain, Are Not Involved in Protein Localization
3.3. The Sequence Linker of AtRMR2 Is Involved in the Protein Traffic to the TGN
3.4. AtRMRs Dimerize in the Secretory Pathway
3.5. AtRMR Traffic and Dimerization
4. Materials and Methods
4.1. Plant Material and Growth Condition
4.2. Agro-Infiltration of N. benthamiana Leaves
4.3. Confocal Microscopy
4.4. Total RNA Extraction and cDNA Synthesis
4.5. Constructions and Plasmids
4.5.1. Vectors for C-Terminal Fusion with Different Fluorescent Reporters
4.5.2. Vectors for N-Terminal Fusion with Different Fluorescent Reporters
4.5.3. Vectors for Bimolecular Fluorescent Complementation (BiFC)
4.5.4. Constructs for the Expression of Full-Length AtRMR1 and -2
4.5.5. Constructs for the Expression of AtRMR2 and 1 Deletion Mutants
4.5.6. Constructs for the Expression of AtRMR2 and 1 Replacement Mutants of the Transmembrane Domain and Sequence Linker
4.5.7. Constructs for the Expression of Protein Markers of Different Compartments
4.5.8. Constructs for Bimolecular Fluorescent Complementation (BiFC)
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 35S | Cauliflower Mosaic Virus (CaMV) promoter |
| aa | Amino acid |
| BiFC | Bimolecular Fluorescence Complementation |
| BP80 | Binding Protein of 80 kDa |
| BYV | Beet Yellow Virus |
| CFP | Cyan Fluorescent Protein |
| cYFP | C-terminal YFP fragment |
| DV | Dense Vesicle |
| eGFP | Enhanced GFP |
| ER | Endoplasmic Reticulum |
| Gly6 | Spacer composed by 6 Glycines |
| GFP | Green Fluorescent Protein |
| GONST1 | Golgi localized GDP-mannose Transporter |
| HA tag | Human Influenza Hemagglutinin (HA) derived protein-tag |
| L | Cytosolic sequence linker from AtRMRs |
| LV | Lytic Vacuole |
| MCS | Multi Cloning Site |
| Myc | Protein-tag derived from the c-myc gene |
| mRFP | Monomeric RFP |
| mCherry | mCherry Fluorescent Protein |
| nYFP | N-terminal YFP fragment |
| p6 | 6 kDa protein from BYV |
| PA | Protein Associated Domain |
| PAC | Precursor Accumulating Vesicle |
| PCR | Polymerase Chain Reaction |
| PSV | Protein Storage Vacuole |
| PVC | Prevacuolar Compartment |
| R | Arginine |
| RING-H2 | RING (Really Interesting New Gene)-H2 domain |
| RFP | Red Fluorescent Protein |
| RMR | Receptor-like Membrane RING-H2 |
| S | Serine |
| ssVSD | Sequence Specific VSD |
| Ser-Rich | Domain Rich in Serines |
| Sp | Signal Peptide |
| SYP61 | Syntaxin protein 61 |
| T | Threonine |
| TGN | Trans-Golgi Network |
| TM | Transmembrane Domain |
| Venus | Venus Fluorescent Protein |
| VSD | Vacuolar Sorting Determinant |
| VSR | Vacuolar Sorting Receptor |
| YFP | Yellow Fluorescent Protein |
References
- Bassham, D.C.; Brandizzi, F.; Otegui, M.S.; Sanderfoot, A.A. The secretory system of Arabidopsis. Arab. Book 2008, 6, e0116. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.G.; Neuhaus, J.M. Receptor-mediated sorting of soluble vacuolar proteins: Myths, facts, and a new model. J. Exp. Bot. 2016. [Google Scholar] [CrossRef] [PubMed]
- Vitale, A.; Raikhel, N.V. What do proteins need to reach different vacuoles? Trends Plant Sci. 1999, 4, 149–155. [Google Scholar] [CrossRef]
- Xiang, L.; Etxeberria, E.; van den Ende, W. Vacuolar protein sorting mechanisms in plants. FEBS J. 2013, 280, 979–993. [Google Scholar] [CrossRef] [PubMed]
- Von Heijne, G. Protein targeting signals. Curr. Opin. Cell Biol. 1990, 2, 604–608. [Google Scholar] [CrossRef]
- Vitale, A.; Ceriotti, A.; Denecke, J. The role of the endoplasmic reticulum in protein synthesis, modification and intracellular transport. J. Exp. Bot. 1993, 44, 1417–1444. [Google Scholar] [CrossRef]
- Boston, R.S.; Viitanen, P.V.; Vierling, E. Molecular chaperones and protein folding in plants. Plant Mol. Biol. 1996, 32, 191–222. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, K.; Neuhaus, J.M. cis-Elements of protein transport to the plant vacuoles. J. Exp. Bot. 1999, 50, 165–174. [Google Scholar] [CrossRef]
- Neuhaus, J.M.; Pietrzak, M.; Boller, T. Mutation analysis of the C-terminal vacuolar targeting peptide of tobacco chitinase: Low specificity of the sorting system, and gradual transition between intracellular retention and secretion into the extracellular space. Plant J. 1994, 5, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Yan, P.K.; Kim, H.; Hwang, I.; Jiang, L. Localization of green fluorescent protein fusions with the seven Arabidopsis vacuolar sorting receptors to prevacuolar compartments in tobacco BY-2 cells. Plant Physiol. 2006, 142, 945–962. [Google Scholar] [CrossRef] [PubMed]
- Hinz, G.; Colanesi, S.; Hillmer, S.; Rogers, J.C.; Robinson, D.G. Localization of vacuolar transport receptors and cargo proteins in the Golgi apparatus of developing Arabidopsis embryos. Traffic 2007, 8, 1452–1464. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, T.; Paris, N.; Butler, J.M.; Beevers, L.; Rogers, J.C. Purification and initial characterization of a potential plant vacuolar targeting receptor. Proc. Natl. Acad. Sci. USA 1994, 91, 3403–3407. [Google Scholar] [CrossRef] [PubMed]
- Paris, N.; Rogers, S.W.; Jiang, L.; Kirsch, T.; Beevers, L.; Phillips, T.E.; Rogers, J.C. Molecular cloning and further characterization of a probable plant vacuolar sorting receptor. Plant Physiol. 1997, 115, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Marty, F. Plant vacuoles. Plant Cell 1999, 11, 587–600. [Google Scholar] [CrossRef] [PubMed]
- Frigerio, L.; Hinz, G.; Robinson, D.G. Multiple vacuoles in plant cells: Rule or exception? Traffic 2008, 9, 1564–1570. [Google Scholar] [CrossRef] [PubMed]
- Jauh, G.Y.; Phillips, T.E.; Rogers, J.C. Tonoplast intrinsic protein isoforms as markers for vacuolar functions. Plant Cell 1999, 11, 1867–1882. [Google Scholar] [CrossRef] [PubMed]
- Di Sansebastiano, G.P.; Paris, N.; Marc-Martin, S.; Neuhaus, J.M. Specific accumulation of GFP in a non-acidic vacuolar compartment via a C-terminal propeptide-mediated sorting pathway. Plant J. 1998, 15, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Rogers, S.W.; Butler, J.; Beevers, L.; Rogers, J.C. Structural requirements for ligand binding by a probable plant vacuolar sorting receptor. Plant Cell 2000, 12, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Fong, Y.H.; Zeng, Y.; Shen, J.; Jiang, L.; Wong, K.B. How vacuolar sorting receptor proteins interact with their cargo proteins: Crystal structures of Apo and cargo-bound forms of the protease-associated domain from an Arabidopsis vacuolar sorting receptor. Plant Cell 2014, 26, 3693–3708. [Google Scholar] [CrossRef] [PubMed]
- Katoh, S.; Tsunoda, Y.; Murata, K.; Minami, E.; Katoh, E. Active site residues and amino acid specificity of the ubiquitin carrier protein-binding RING-H2 finger domain. J. Biol. Chem. 2005, 280, 41015–41024. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Phillips, T.E.; Rogers, S.W.; Rogers, J.C. Biogenesis of the protein storage vacuole crystalloid. J. Cell Biol. 2000, 150, 755–770. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, T.; Saalbach, G.; Raikhel, N.V.; Beevers, L. Interaction of a potential vacuolar targeting receptor with amino- and carboxyl-terminal targeting determinants. Plant Physiol. 1996, 111, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.U.; Rojo, E.; Kovaleva, V.; Venkataraman, S.; Dombrowski, J.E.; Matsuoka, K.; Raikhel, N.V. The plant vacuolar sorting receptor AtELP is involved in transport of NH2-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J. Cell Biol. 2000, 149, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Lee, D.; Lee, G.J.; Hwang, I. AtRMR1 functions as a cargo receptor for protein trafficking to the protein storage vacuole. J. Cell Biol. 2005, 170, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Oufattole, M.; Rogers, J.C. Golgi-mediated vacuolar sorting in plant cells: RMR proteins are sorting receptors for the protein aggregation/membrane internalization pathway. Plant Sci. 2007, 172, 728–745. [Google Scholar] [CrossRef]
- Scabone, C.M.; Frigerio, L.; Petruccelli, S. A fluorescent reporter protein containing AtRMR1 domains is targeted to the storage and central vacuoles in Arabidopsis thaliana and tobacco leaf cells. Plant Cell Rep. 2011, 30, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Peremyslov, V.V.; Pan, Y.W.; Dolja, V.V. Movement protein of a closterovirus is a type III integral transmembrane protein localized to the endoplasmic reticulum. J. Virol. 2004, 78, 3704–3709. [Google Scholar] [CrossRef] [PubMed]
- Uemura, T.; Ueda, T.; Ohniwa, R.L.; Nakano, A.; Takeyasu, K.; Sato, M.H. Systematic analysis of SNARE molecules in Arabidopsis: Dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 2004, 29, 49–65. [Google Scholar] [CrossRef] [PubMed]
- Foresti, O.; Denecke, J. Intermediate organelles of the plant secretory pathway: Identity and function. Traffic 2008, 9, 1599–1612. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kang, H.; Jang, M.; Chang, J.H.; Miao, Y.; Jiang, L.; Hwang, I. Homomeric interaction of AtVSR1 is essential for its function as a vacuolar sorting receptor. Plant Physiol. 2010, 154, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Ohad, N.; Shichrur, K.; Yalovsky, S. The analysis of protein–protein interactions in plants by bimolecular fluorescence complementation. Plant Physiol. 2007, 145, 1090–1099. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, I.; Hamilton, A.D.; Regan, L. Antiparallel leucine zipper-directed protein reassembly: Application to the green fluorescent protein. J. Am. Chem. Soc. 2000, 122, 5658–5659. [Google Scholar] [CrossRef]
- Virgili-Lopez, G.; Langhans, M.; Bubeck, J.; Pedrazzini, E.; Gouzerh, G.; Neuhaus, J.M.; Robinson, D.G.; Vitale, A. Comparison of membrane targeting strategies for the accumulation of the human immunodeficiency virus p24 protein in transgenic tobacco. Int. J. Mol. Sci. 2013, 14, 13241–13265. [Google Scholar] [CrossRef] [PubMed]
- Virgili-Lopez, G. Targeting to Compartments of the Endomembrane System for the Accumulation of HIV-1 p24 in Tobacco Plants. Ph.D. Thesis, Ruprecht-Karls-Universität, Heidelberg, Germany, 2008. [Google Scholar]
- Van Dijk, J.R.; Yamazaki, Y.; Palmer, R.H. Tumour-associated mutations of PA-TM-RING ubiquitin ligases RNF167/RNF13 identify the PA domain as a determinant for endosomal localization. Biochem. J. 2014, 459, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Brandizzi, F.; Frangne, N.; Marc-Martin, S.; Hawes, C.; Neuhaus, J.M.; Paris, N. The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 2002, 14, 1077–1092. [Google Scholar] [CrossRef] [PubMed]
- Happel, N.; Honing, S.; Neuhaus, J.M.; Paris, N.; Robinson, D.G.; Holstein, S.E. Arabidopsis µA-adaptin interacts with the tyrosine motif of the vacuolar sorting receptor VSR-PS1. Plant J. 2004, 37, 678–693. [Google Scholar] [CrossRef] [PubMed]
- Saint-Jean, B.; Seveno-Carpentier, E.; Alcon, C.; Neuhaus, J.M.; Paris, N. The cytosolic tail dipeptide Ile-Met of the pea receptor BP80 is required for recycling from the prevacuole and for endocytosis. Plant Cell 2010, 22, 2825–2837. [Google Scholar] [CrossRef] [PubMed]
- Giraudo, C.G.; Maccioni, H.J. Endoplasmic reticulum export of glycosyltransferases depends on interaction of a cytoplasmic dibasic motif with Sar1. Mol. Biol. Cell 2003, 14, 3753–3766. [Google Scholar] [CrossRef] [PubMed]
- Schoberer, J.; Vavra, U.; Stadlmann, J.; Hawes, C.; Mach, L.; Steinkellner, H.; Strasser, R. Arginine/lysine residues in the cytoplasmic tail promote ER export of plant glycosylation enzymes. Traffic 2009, 10, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Jurgens, G.; Geldner, N. The high road and the low road: Trafficking choices in plants. Cell 2007, 130, 977–979. [Google Scholar] [CrossRef] [PubMed]
- Mahon, P.; Bateman, A. The PA domain: A protease-associated domain. Protein Sci. 2000, 9, 1930–1934. [Google Scholar] [CrossRef] [PubMed]
- D’Azzo, A.; Bongiovanni, A.; Nastasi, T. E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic 2005, 6, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Wimley, W.C.; Hristova, K. Transmembrane helix dimerization: Beyond the search for sequence motifs. Biochim. Biophys. Acta 2012, 1818, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Stigliano, E.; Faraco, M.; Neuhaus, J.M.; Montefusco, A.; Dalessandro, G.; Piro, G.; di Sansebastiano, G.P. Two glycosylated vacuolar GFPs are new markers for ER-to-vacuole sorting. Plant Physiol. Biochem. 2013, 73, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Stigliano, E.; di Sansebastiano, G.P.; Neuhaus, J.M. Contribution of chitinase A’s C-terminal vacuolar sorting determinant to the study of soluble protein compartmentation. Int. J. Mol. Sci. 2014, 15, 11030–11039. [Google Scholar] [CrossRef] [PubMed]
- Sanmartin, M.; Ordonez, A.; Sohn, E.J.; Robert, S.; Sanchez-Serrano, J.J.; Surpin, M.A.; Raikhel, N.V.; Rojo, E. Divergent functions of VTI12 and VTI11 in trafficking to storage and lytic vacuoles in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 3645–3650. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Rogers, J.C.; Jiang, L. Plant RMR proteins: Unique vacuolar sorting receptors that couple ligand sorting with membrane internalization. FEBS J. 2011, 278, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Uemura, T.; Suda, Y.; Ueda, T.; Nakano, A. Dynamic behavior of the trans-Golgi network in root tissues of Arabidopsis revealed by super-resolution live imaging. Plant Cell Physiol. 2014, 55, 694–703. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.T.; Occhialini, A.; Andralojc, P.J.; Devonshire, J.; Hines, K.M.; Parry, M.A.; Hanson, M.R. β-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J. 2014, 79, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O.; Rivas, S.; Mestre, P.; Baulcombe, D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003, 33, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Hellens, R.P.; Edwards, E.A.; Leyland, N.R.; Bean, S.; Mullineaux, P.M. pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000, 42, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, J.M.; Ahl-Goy, P.; Hinz, U.; Flores, S.; Meins, F., Jr. High-level expression of a tobacco chitinase gene in Nicotiana sylvestris. Susceptibility of transgenic plants to Cercospora nicotianae infection. Plant Mol. Biol. 1991, 16, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, T.C.; Handford, M.G.; Yuseff, M.I.; Orellana, A.; Dupree, P. Identification and characterization of GONST1, a Golgi-localized GDP-mannose transporter in Arabidopsis. Plant Cell 2001, 13, 2283–2295. [Google Scholar] [CrossRef] [PubMed]










© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Occhialini, A.; Gouzerh, G.; Di Sansebastiano, G.-P.; Neuhaus, J.-M. Dimerization of the Vacuolar Receptors AtRMR1 and -2 from Arabidopsis thaliana Contributes to Their Localization in the trans-Golgi Network. Int. J. Mol. Sci. 2016, 17, 1661. https://doi.org/10.3390/ijms17101661
Occhialini A, Gouzerh G, Di Sansebastiano G-P, Neuhaus J-M. Dimerization of the Vacuolar Receptors AtRMR1 and -2 from Arabidopsis thaliana Contributes to Their Localization in the trans-Golgi Network. International Journal of Molecular Sciences. 2016; 17(10):1661. https://doi.org/10.3390/ijms17101661
Chicago/Turabian StyleOcchialini, Alessandro, Guillaume Gouzerh, Gian-Pietro Di Sansebastiano, and Jean-Marc Neuhaus. 2016. "Dimerization of the Vacuolar Receptors AtRMR1 and -2 from Arabidopsis thaliana Contributes to Their Localization in the trans-Golgi Network" International Journal of Molecular Sciences 17, no. 10: 1661. https://doi.org/10.3390/ijms17101661
APA StyleOcchialini, A., Gouzerh, G., Di Sansebastiano, G. -P., & Neuhaus, J. -M. (2016). Dimerization of the Vacuolar Receptors AtRMR1 and -2 from Arabidopsis thaliana Contributes to Their Localization in the trans-Golgi Network. International Journal of Molecular Sciences, 17(10), 1661. https://doi.org/10.3390/ijms17101661