Effects of Glycosylation on the Enzymatic Activity and Mechanisms of Proteases
Abstract
:1. Introduction
2. Glycosylated Proteases
2.1. Aspartic Proteases
2.2. Cysteine Proteases
2.3. Metalloproteases
2.4. Serine Proteases
2.5. Threonine Proteases
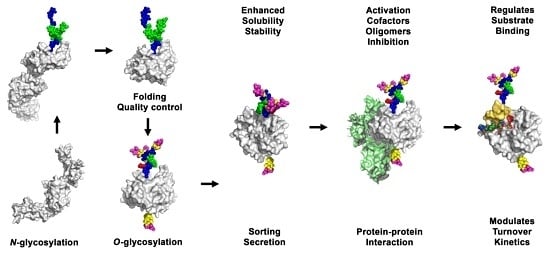
3. Effects of Glycosylation on Proteases
3.1. Effects of Glycosylation on Folding, Sub-Cellular Distribution and Secretion of Proteases
3.2. Effects of Glycosylation on Activation and Stability of Proteases
3.3. Effects of Glycosylation on Substrate Binding and Turnover
3.4. Effects of Glycosylation on Protease Structures
3.5. Effects of Glycosylation on Protease Mechanisms
4. Conclusions and Outlook
Acknowledgments
Conflicts of Interest
References
- Dell, A.; Galadari, A.; Sastre, F.; Hitchen, P. Similarities and Differences in the Glycosylation Mechanisms in Prokaryotes and Eukaryotes. Int. J. Microbiol. 2010, 2010, 148178. [Google Scholar] [CrossRef] [PubMed]
- Apweiler, R.; Hermjakob, H.; Sharon, N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1999, 1473, 4–8. [Google Scholar] [CrossRef]
- Varki, A.; Cummings, R.D.; Esko, J.D.; Freeze, H.H.; Stanley, P.; Bertozzi, C.R.; Hart, G.W.; Etzler, M.E. Essentials of Glycobiology, 2nd ed.; Varki, A., Cummings, R.D., Esko, J.D., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009. [Google Scholar]
- Mitra, N.; Sinha, S.; Ramya, T.N.C.; Surolia, A. N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem. Sci. 2006, 31, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.; Oldham, N.J.; Davis, B.G. Site-selective chemical protein glycosylation protects from autolysis and proteolytic degradation. Carbohydr. Res. 2009, 344, 1508–1514. [Google Scholar] [CrossRef] [PubMed]
- Kozarsky, K.; Kingsley, D.; Krieger, M. Use of a mutant cell line to study the kinetics and function of O-linked glycosylation of low density lipoprotein receptors. Proc. Natl. Acad. Sci. USA 1988, 85, 4335–4339. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.; Krieger, M. Isolation and characterization of an extragenic suppressor of the low-density lipoprotein receptor-deficient phenotype of a Chinese hamster ovary cell mutant. Mol. Cell. Biol. 1989, 9, 4799–4806. [Google Scholar] [CrossRef] [PubMed]
- Schjoldager, K.T.B.G.; Clausen, H. Site-specific protein O-glycosylation modulates proprotein processing—Deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochim. Biophys. Acta 2012, 1820, 2079–2094. [Google Scholar] [CrossRef] [PubMed]
- Moremen, K.W.; Tiemeyer, M.; Nairn, A.V. Vertebrate protein glycosylation: Diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 2012, 13, 448–462. [Google Scholar] [CrossRef] [PubMed]
- Rabinovich, G.A.; van Kooyk, Y.; Cobb, B.A. Glycobiology of immune responses. Ann. N. Y. Acad. Sci. 2012, 1253, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, K.; Marth, J.D. Glycosylation in Cellular Mechanisms of Health and Disease. Cell 2006, 126, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hagen, T.; Kelly, G. The cellular microenvironment and cell adhesion: A role for O-glycosylation. Biochem. Soc. Trans. 2011, 39, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Martínek, V.; Sklenář, J.; Dračínský, M.; Šulc, M.; Hofbauerová, K.; Bezouška, K.; Frei, E.; Stiborová, M. Glycosylation Protects Proteins against Free Radicals Generated from Toxic Xenobiotics. Toxicol. Sci. 2010, 117, 359–374. [Google Scholar] [CrossRef] [PubMed]
- Zafar, S.; Nasir, A.; Bokhari, H. Computational analysis reveals abundance of potential glycoproteins in Archaea, Bacteria and Eukarya. Bioinformation 2011, 6, 352–355. [Google Scholar] [CrossRef] [PubMed]
- Gavel, Y.; Heijne, G. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: Implications for protein engineering. Protein Eng. 1990, 3, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Zielinska, D.F.; Gnad, F.; Wiśniewski, J.R.; Mann, M. Precision Mapping of an In Vivo N-Glycoproteome Reveals Rigid Topological and Sequence Constraints. Cell 2010, 141, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Stanley, P.; Schachter, H.; Taniguchi, N. Chapter 8: N-Glycans. In Essentials of Glycobiology, 2nd ed.; Varki, A., Cummings, R.D., Esko, J.D., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009. [Google Scholar]
- Nyathi, Y.; Wilkinson, B.M.; Pool, M.R. Co-translational targeting and translocation of proteins to the endoplasmic reticulum. Biochim. Biophys. Acta 2013, 1833, 2392–2402. [Google Scholar] [CrossRef] [PubMed]
- Akopian, D.; Shen, K.; Zhang, X.; Shan, S.-O. Signal Recognition Particle: An Essential Protein-Targeting Machine. Annu. Rev. Biochem. 2013, 82, 693–721. [Google Scholar] [CrossRef] [PubMed]
- Aebi, M. N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 2013, 1833, 2430–2437. [Google Scholar] [CrossRef] [PubMed]
- Oka, O.B.V.; Bulleid, N.J. Forming disulfides in the endoplasmic reticulum. Biochim. Biophys. Acta 2013, 1833, 2425–2429. [Google Scholar] [CrossRef] [PubMed]
- Gidalevitz, T.; Stevens, F.; Argon, Y. Orchestration of secretory protein folding by ER chaperones. Biochim. Biophys. Acta 2013, 1833, 2410–2424. [Google Scholar] [CrossRef] [PubMed]
- Appenzeller-Herzog, C.; Hauri, H.-P. The ER-Golgi intermediate compartment (ERGIC): In search of its identity and function. J. Cell Sci. 2006, 119, 2173–2183. [Google Scholar] [CrossRef] [PubMed]
- Anelli, T.; Sitia, R. Protein quality control in the early secretory pathway. EMBO J. 2008, 27, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Kellokumpu, S.; Hassinen, A.; Glumoff, T. Glycosyltransferase complexes in eukaryotes: Long-known, prevalent but still unrecognized. Cell. Mol. Life Sci. 2016, 73, 305–325. [Google Scholar] [CrossRef] [PubMed]
- Kizuka, Y.; Taniguchi, N. Enzymes for N-Glycan Branching and Their Genetic and Nongenetic Regulation in Cancer. Biomolecules 2016, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Wopereis, S.; Lefeber, D.J.; Morava, É.; Wevers, R.A. Mechanisms in Protein O-Glycan Biosynthesis and Clinical and Molecular Aspects of Protein O-Glycan Biosynthesis Defects: A Review. Clin. Chem. 2006, 52, 574–600. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.V.N.; Goldman, R.; Karagiannis, K.; Narsule, T.; Simonyan, V.; Soika, V.; Mazumder, R. Structure-based Comparative Analysis and Prediction of N-linked Glycosylation Sites in Evolutionarily Distant Eukaryotes. Genom. Proteom. Bioinform. 2013, 11, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Brunak, S. Prediction of glycosylation across the human proteome and the correlation to protein function. In Pacific Symposium on Biocomputing; World Scientific Press: Lihue, HI, USA, 2002; pp. 310–322. [Google Scholar]
- Zielinska, D.F.; Gnad, F.; Schropp, K.; Wiśniewski Jacek, R.; Mann, M. Mapping N-Glycosylation Sites across Seven Evolutionarily Distant Species Reveals a Divergent Substrate Proteome Despite a Common Core Machinery. Mol. Cell 2012, 46, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Christlet, T.H.T.; Veluraja, K. Database Analysis of O-Glycosylation Sites in Proteins. Biophys. J. 2001, 80, 952–960. [Google Scholar] [CrossRef]
- Hansen, J.E.; Lund, O.; Nielsen, J.O.; Hansen, J.-E.S.; Brunak, S. O-GLYCBASE: A Revised Database of O-Glycosylated Proteins. Nucleic Acids Res. 1996, 24, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Steen, P.V.; Rudd, P.M.; Dwek, R.A.; Opdenakker, G. Concepts and Principles of O-Linked Glycosylation. Crit. Rev. Biochem. Mol. Biol. 1998, 33, 151–208. [Google Scholar] [CrossRef] [PubMed]
- Tian, E.; Ten Hagen, K.G. Recent insights into the biological roles of mucin-type O-glycosylation. Glycoconj. J. 2009, 26, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Lommel, M.; Strahl, S. Protein O-mannosylation: Conserved from bacteria to humans. Glycobiology 2009, 19, 816–828. [Google Scholar] [CrossRef] [PubMed]
- Song, E.; Mechref, Y. LC–MS/MS Identification of the O-Glycosylation and Hydroxylation of Amino Acid Residues of Collagen α-1 (II) chain from Bovine Cartilage. J. Proteome Res. 2013, 12, 3599–3609. [Google Scholar] [CrossRef] [PubMed]
- Hijazi, M.; Velasquez, M.S.; Jamet, E.; Estevez, J.M.; Albenne, C. An update on post-translational modifications of hydroxyproline-rich glycoproteins: Towards a model highlighting their contribution to plant cell wall architecture. Front. Plant Sci. 2014, 5, 395. [Google Scholar] [CrossRef] [PubMed]
- Hofsteenge, J.; Blommers, M.; Hess, D.; Furmanek, A.; Miroshnichenko, O. The Four Terminal Components of the Complement System AreC-Mannosylated on Multiple Tryptophan Residues. J. Biol. Chem. 1999, 274, 32786–32794. [Google Scholar] [CrossRef] [PubMed]
- Ihara, Y.; Inai, Y.; Ikezaki, M.; Matsui, I.-S.L.; Manabe, S.; Ito, Y. C-Mannosylation: A Modification on Tryptophan in Cellular Proteins. In Glycoscience: Biology and Medicine; Endo, T., Seeberger, H.P., Hart, W.G., Wong, C.-H., Taniguchi, N., Eds.; Springer: Tokyo, Japan, 2015; pp. 1–8. [Google Scholar]
- Zurzolo, C.; Simons, K. Glycosylphosphatidylinositol-anchored proteins: Membrane organization and transport. Biochim. Biophys. Acta 2016, 1858, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Nedić, O.; Rattan, S.I.S.; Grune, T.; Trougakos, I.P. Molecular effects of advanced glycation end products on cell signaling pathways, ageing and pathophysiology. Free Radic. Res. 2013, 47, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Henning, C.; Glomb, M.A. Pathways of the Maillard reaction under physiological conditions. Glycoconj. J. 2016, 33, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.U. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef] [PubMed]
- Julenius, K.; Mølgaard, A.; Gupta, R.; Brunak, S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 2005, 15, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Barrett, A.J.; Bateman, A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012, 40, D343–D350. [Google Scholar] [CrossRef] [PubMed]
- Schechter, I.; Berger, A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967, 27, 157–162. [Google Scholar] [CrossRef]
- Ben Halima, S.; Mishra, S.; Raja, K.; Muruga, P.; Willem, M.; Baici, A.; Simons, K.; Brüstle, O.; Koch, P.; Haass, C.; et al. Specific Inhibition of β-Secretase Processing of the Alzheimer Disease Amyloid Precursor Protein. Cell Rep. 2016, 14, 2127–2141. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Reichert, P.; Beyer, B.M.; Liu, J.; Lee, J.; Zhang, L.; Liu, Y.-H.; Taremi, S.S.; Le, H.V.; Strickland, C. Crystallization of glycosylated human BACE protease domain expressed in Trichoplusia ni. Biochim. Biophys. Acta 2004, 1698, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Novinec, M.; Lenar, B.; Turk, B. Cysteine Cathepsin Activity Regulation by Glycosaminoglycans. BioMed Res. Int. 2014, 2014, 309718. [Google Scholar] [CrossRef] [PubMed]
- McLuskey, K.; Mottram, J.C. Comparative structural analysis of the caspase family with other clan CD cysteine peptidases. Biochem. J. 2015, 466, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Campbell, R.L.; Davies, P.L. Structure–function relationships in calpains. Biochem. J. 2012, 447, 335–351. [Google Scholar] [CrossRef] [PubMed]
- Dall, E.; Brandstetter, H. Mechanistic and structural studies on legumain explain its zymogenicity, distinct activation pathways, and regulation. Proc. Natl. Acad. Sci. USA 2013, 110, 10940–10945. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-M.; Dando, P.M.; Rawlings, N.D.; Brown, M.A.; Young, N.E.; Stevens, R.A.; Hewitt, E.; Watts, C.; Barrett, A.J. Cloning, Isolation, and Characterization of Mammalian Legumain, an Asparaginyl Endopeptidase. J. Biol. Chem. 1997, 272, 8090–8098. [Google Scholar] [CrossRef] [PubMed]
- Klein, T.; Bischoff, R. Physiology and pathophysiology of matrix metalloproteases. Amino Acids 2011, 41, 271–290. [Google Scholar] [CrossRef] [PubMed]
- Bertini, I.; Calderone, V.; Fragai, M.; Jaiswal, R.; Luchinat, C.; Melikian, M.; Mylonas, E.; Svergun, D.I. Evidence of Reciprocal Reorientation of the Catalytic and Hemopexin-Like Domains of Full-Length MMP-12. J. Am. Chem. Soc. 2008, 130, 7011–7021. [Google Scholar] [CrossRef] [PubMed]
- Boon, L.; Ugarte-Berzal, E.; Vandooren, J.; Opdenakker, G. Glycosylation of matrix metalloproteases and tissue inhibitors: Present state, challenges and opportunities. Biochem. J. 2016, 473, 1471–1482. [Google Scholar] [CrossRef] [PubMed]
- Duellman, T.; Burnett, J.; Yang, J. Functional Roles of N-Linked Glycosylation of Human Matrix Metalloproteinase 9. Traffic 2015, 16, 1108–1126. [Google Scholar] [CrossRef] [PubMed]
- Mattu, T.S.; Royle, L.; Langridge, J.; Wormald, M.R.; van den Steen, P.E.; van Damme, J.; Opdenakker, G.; Harvey, D.J.; Dwek, R.A.; Rudd, P.M. O-Glycan Analysis of Natural Human Neutrophil Gelatinase B Using a Combination of Normal Phase-HPLC and Online Tandem Mass Spectrometry: Implications for the Domain Organization of the Enzyme. Biochemistry 2000, 39, 15695–15704. [Google Scholar] [CrossRef] [PubMed]
- Fry, S.A.; van den Steen, P.E.; Royle, L.; Wormald, M.R.; Leathem, A.J.; Opdenakker, G.; McDonnell, J.M.; Dwek, R.A.; Rudd, P.M. Cancer-Associated Glycoforms of Gelatinase B Exhibit a Decreased Level of Binding to Galectin-3. Biochemistry 2006, 45, 15249–15258. [Google Scholar] [CrossRef] [PubMed]
- Seals, D.F.; Courtneidge, S.A. The ADAMs family of metalloproteases: Multidomain proteins with multiple functions. Genes Dev. 2003, 17, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Kelwick, R.; Desanlis, I.; Wheeler, G.N.; Edwards, D.R. The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol. 2015, 16, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goth, C.K.; Halim, A.; Khetarpal, S.A.; Rader, D.J.; Clausen, H.; Schjoldager, K.T.-B.G. A systematic study of modulation of ADAM-mediated ectodomain shedding by site-specific O-glycosylation. Proc. Natl. Acad. Sci. USA 2015, 112, 14623–14628. [Google Scholar] [CrossRef] [PubMed]
- Arolas, J.L.; Broder, C.; Jefferson, T.; Guevara, T.; Sterchi, E.E.; Bode, W.; Stöcker, W.; Becker-Pauly, C.; Gomis-Rüth, F.X. Structural basis for the sheddase function of human meprin β metalloproteinase at the plasma membrane. Proc. Natl. Acad. Sci. USA 2012, 109, 16131–16136. [Google Scholar] [CrossRef] [PubMed]
- Broder, C.; Becker-Pauly, C. The metalloproteases meprin α and meprin β: Unique enzymes in inflammation, neurodegeneration, cancer and fibrosis. Biochem. J. 2013, 450, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Bertenshaw, G.P.; Norcum, M.T.; Bond, J.S. Structure of Homo- and Hetero-oligomeric Meprin Metalloproteases: Dimers, tetramers, and high molecular mass multimers. J. Biol. Chem. 2003, 278, 2522–2532. [Google Scholar] [CrossRef] [PubMed]
- Fernández, D.; Pallarès, I.; Vendrell, J.; Avilés, F.X. Progress in metallocarboxypeptidases and their small molecular weight inhibitors. Biochimie 2010, 92, 1484–1500. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-Y.; Rong, Y.; Correia, K.; Min, J.; Morgan, J.I. Comparison of the Enzymatic and Functional Properties of Three Cytosolic Carboxypeptidase Family Members. J. Biol. Chem. 2015, 290, 1222–1232. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; MacDonald, B.T.; Gao, H.; Shamashkin, M.; Coyle, A.J.; Martinez, R.V.; He, X. Characterization of Tiki, a New Family of Wnt-specific Metalloproteases. J. Biol. Chem. 2016, 291, 2435–2443. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.L.; Kitamoto, Y.; Sadler, J.E. Enteropeptidase, a type II transmembrane serine protease. Front. Biosci. 2009, 1, 242–249. [Google Scholar]
- Smith, S.A.; Travers, R.J.; Morrissey, J.H. How it all starts: Initiation of the clotting cascade. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Johansson, L.; Karpf, D.M.; Hansen, L.; Pelzer, H.; Persson, E. Activation peptides prolong the murine plasma half-life of human factor VII. Blood 2011, 117, 3445–3452. [Google Scholar] [CrossRef] [PubMed]
- Begbie, M.E.; Mamdani, A.; Gataiance, S.; Eltringham-Smith, L.J.; Bhakta, V.; Hortelano, G.; Sheffield, W.P. An important role for the activation peptide domain in controlling factor IX levels in the blood of haemophilia B mice. Thromb. Haemost. 2005, 94, 1138–1147. [Google Scholar] [CrossRef] [PubMed]
- Aisina, R.; Mukhametova, L.; Gershkovich, K.; Varfolomeyev, S. The role of carbohydrate side chains of plasminogen in its activation by staphylokinase. Biochim. Biophys. Acta 2005, 1725, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Law, R.H.P.; Caradoc-Davies, T.; Cowieson, N.; Horvath, A.J.; Quek, A.J.; Encarnacao, J.A.; Steer, D.; Cowan, A.; Zhang, Q.; Lu, B.G.C.; et al. The X-ray Crystal Structure of Full-Length Human Plasminogen. Cell Rep. 2012, 1, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Buko, A.M.; Kentzer, E.J.; Petros, A.; Menon, G.; Zuiderweg, E.R.; Sarin, V.K. Characterization of a posttranslational fucosylation in the growth factor domain of urinary plasminogen activator. Proc. Natl. Acad. Sci. USA 1991, 88, 3992–3996. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.J.; Leonard, C.K.; Guzzetta, A.W.; Spellman, M.W. Tissue plasminogen activator has an O-linked fucose attached to threonine-61 in the epidermal growth factor domain. Biochemistry 1991, 30, 2311–2314. [Google Scholar] [CrossRef] [PubMed]
- Lundwall, A.; Brattsand, M. Kallikrein-related peptidases. Cell. Mol. Life Sci. 2008, 65, 2019–2038. [Google Scholar] [CrossRef] [PubMed]
- Prassas, I.; Eissa, A.; Poda, G.; Diamandis, E.P. Unleashing the therapeutic potential of human kallikrein-related serine proteases. Nat. Rev. Drug Discov. 2015, 14, 183–202. [Google Scholar] [CrossRef] [PubMed]
- Sotiropoulou, G.; Pampalakis, G. Targeting the kallikrein-related peptidases for drug development. Trends Pharmacol. Sci. 2012, 33, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Skala, W.; Magdolen, V.; Brandstetter, H.; Goettig, P. Sweetened kallikrein-related peptidases (KLKs): Glycan trees as potential regulators of activation and activity. Biol. Chem. 2014, 395, 959–976. [Google Scholar] [CrossRef] [PubMed]
- Kalinska, M.; Meyer-Hoffert, U.; Kantyka, T.; Potempa, J. Kallikreins—The melting pot of activity and function. Biochimie 2016, 122, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Seidah, N.G.; Sadr, M.S.; Chrétien, M.; Mbikay, M. The Multifaceted Proprotein Convertases: Their Unique, Redundant, Complementary, and Opposite Functions. J. Biol. Chem. 2013, 288, 21473–21481. [Google Scholar] [CrossRef] [PubMed]
- Gram Schjoldager, K.T.-B.; Vester-Christensen, M.B.; Goth, C.K.; Petersen, T.N.; Brunak, S.; Bennett, E.P.; Levery, S.B.; Clausen, H. A Systematic Study of Site-specific GalNAc-type O-Glycosylation Modulating Proprotein Convertase Processing. J. Biol. Chem. 2011, 286, 40122–40132. [Google Scholar] [CrossRef] [PubMed]
- Wujek, P.; Kida, E.; Walus, M.; Wisniewski, K.E.; Golabek, A.A. N-Glycosylation Is Crucial for Folding, Trafficking, and Stability of Human Tripeptidyl-peptidase 1. J. Biol. Chem. 2004, 279, 12827–12839. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Kraetzner, R.; Gruene, T.; Grapp, M.; Schreiber, K.; Grønborg, M.; Urlaub, H.; Becker, S.; Asif, A.R.; Gärtner, J.; et al. Structure of Tripeptidyl-peptidase I Provides Insight into the Molecular Basis of Late Infantile Neuronal Ceroid Lipofuscinosis. J. Biol. Chem. 2009, 284, 3976–3984. [Google Scholar] [CrossRef] [PubMed]
- Engel, M.; Hoffmann, T.; Wagner, L.; Wermann, M.; Heiser, U.; Kiefersauer, R.; Huber, R.; Bode, W.; Demuth, H.-U.; Brandstetter, H. The crystal structure of dipeptidyl peptidase IV (CD26) reveals its functional regulation and enzymatic mechanism. Proc. Natl. Acad. Sci. USA 2003, 100, 5063–5068. [Google Scholar] [CrossRef] [PubMed]
- Aertgeerts, K.; Levin, I.; Shi, L.; Snell, G.P.; Jennings, A.; Prasad, G.S.; Zhang, Y.; Kraus, M.L.; Salakian, S.; Sridhar, V.; et al. Structural and Kinetic Analysis of the Substrate Specificity of Human Fibroblast Activation Protein α. J. Biol. Chem. 2005, 280, 19441–19444. [Google Scholar] [CrossRef] [PubMed]
- Aertgeerts, K.; Ye, S.; Shi, L.; Prasad, S.G.; Witmer, D.; Chi, E.; Sang, B.-C.; Wijnands, R.A.; Webb, D.R.; Swanson, R.V. N-linked glycosylation of dipeptidyl peptidase IV (CD26): Effects on enzyme activity, homodimer formation, and adenosine deaminase binding. Protein Sci. 2004, 13, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Ditzel, L.; Löwe, J.; Stock, D.; Bochtler, M.; Bartunik, H.D.; Huber, R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 1997, 386, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Huber, E.M.; Basler, M.; Schwab, R.; Heinemeyer, W.; Kirk, C.J.; Groettrup, M.; Groll, M. Immuno- and Constitutive Proteasome Crystal Structures Reveal Differences in Substrate and Inhibitor Specificity. Cell 2012, 148, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Van den Eynde, B.t.J.; Morel, S. Differential processing of class-I-restricted epitopes by the standard proteasome and the immunoproteasome. Curr. Opin. Immunol. 2001, 13, 147–153. [Google Scholar] [CrossRef]
- Kikuchi, J.; Iwafune, Y.; Akiyama, T.; Okayama, A.; Nakamura, H.; Arakawa, N.; Kimura, Y.; Hirano, H. Co- and post-translational modifications of the 26S proteasome in yeast. Proteomics 2010, 10, 2769–2779. [Google Scholar] [CrossRef] [PubMed]
- Overath, T.; Kuckelkorn, U.; Henklein, P.; Strehl, B.; Bonar, D.; Kloss, A.; Siele, D.; Kloetzel, P.-M.; Janek, K. Mapping of O-GlcNAc Sites of 20 S Proteasome Subunits and Hsp90 by a Novel Biotin-Cystamine Tag. Mol. Cell. Proteom. 2012, 11, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Solá, R.J.; Griebenow, K. Effects of glycosylation on the stability of protein pharmaceuticals. J. Pharm. Sci. 2009, 98, 1223–1245. [Google Scholar] [CrossRef] [PubMed]
- Bence, M.; Sahin-Toth, M. Asparagine-linked glycosylation of human chymotrypsin C is required for folding and secretion but not for enzyme activity. FEBS J. 2011, 278, 4338–4350. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Wang, X.; Ding, H.; Jin, M.; Yu, L.; Wang, J.; Yu, X. The role of N-glycosylation sites in the activity, stability, and expression of the recombinant elastase expressed by Pichia pastoris. Enzym. Microb. Technol. 2014, 54, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Thayer, M.M.; Flaherty, K.M.; McKay, D.B. Three-dimensional structure of the elastase of Pseudomonas aeruginosa at 1.5-A resolution. J. Biol. Chem. 1991, 266, 2864–2871. [Google Scholar] [PubMed]
- Debela, M.; Goettig, P.; Magdolen, V.; Huber, R.; Schechter, N.M.; Bode, W. Structural Basis of the Zinc Inhibition of Human Tissue Kallikrein 5. J. Mol. Biol. 2007, 373, 1017–1031. [Google Scholar] [CrossRef] [PubMed]
- Koo, B.-H.; Longpré, J.-M.; Somerville, R.P.T.; Alexander, J.P.; Leduc, R.; Apte, S.S. Regulation of ADAMTS9 Secretion and Enzymatic Activity by Its Propeptide. J. Biol. Chem. 2007, 282, 16146–16154. [Google Scholar] [CrossRef] [PubMed]
- Hori, H.; Yoshino, T.; Ishizuka, Y.; Yamauchi, T.; Shiratori, Y.; Nakagawa, S.; Umeyama, H.; Murakami, K. Characterization of N-linked oligosaccharides attached to human renin expressed in COS cells. Clin. Exp. Hypertens. A 1988, 10, 1147–1155. [Google Scholar] [PubMed]
- Kim, S.; Hiruma, M.; Ikemoto, F.; Yamamoto, K. Importance of glycosylation for hepatic clearance of renal renin. Am. J. Physiol. Endocrinol. Metab. 1988, 255, E642–E651. [Google Scholar]
- Dumez, M.-E.; Teller, N.; Mercier, F.; Tanaka, T.; Vandenberghe, I.; Vandenbranden, M.; Devreese, B.; Luxen, A.; Frère, J.-M.; Matagne, A.; et al. Activation Mechanism of Recombinant Der p 3 Allergen Zymogen: Contribution of Cysteine Protease Der p 1 and Effect of Propeptide Glycosylation. J. Biol. Chem. 2008, 283, 30606–30617. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Wang, W.; Chen, S.; Wu, Q. Role of Glycosylation in Corin Zymogen Activation. J. Biol. Chem. 2007, 282, 27728–27735. [Google Scholar] [CrossRef] [PubMed]
- Mach, L.; Stüwe, K.; Hagen, A.; Ballaun, C.; Glössl, J. Proteolytic processing and glycosylation of cathepsin B. The role of the primary structure of the latent precursor and of the carbohydrate moiety for cell-type-specific molecular forms of the enzyme. Biochem. J. 1992, 282, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, Y.; Ikeda, S.; Sakai, H.; Tsukuba, T.; Okamoto, K.; Nishishita, K.; Akamine, A.; Kato, Y.; Yamamoto, K. Role of N-glycosylation in cathepsin E. Eur. J. Biochem. 1999, 266, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Takao, K.; Takai, S.; Shiota, N.; Song, K.; Nishimura, K.; Ishihara, T.; Miyazaki, M. Lack of effect of carbohydrate depletion on some properties of human mast cell chymase. Biochim. Biophys. Acta 1999, 1427, 74–81. [Google Scholar] [CrossRef]
- Ihara, S.; Miyoshi, E.; Nakahara, S.; Sakiyama, H.; Ihara, H.; Akinaga, A.; Honke, K.; Dickson, R.B.; Lin, C.-Y.; Taniguchi, N. Addition of β1–6 GlcNAc branching to the oligosaccharide attached to Asn 772 in the serine protease domain of matriptase plays a pivotal role in its stability and resistance against trypsin. Glycobiology 2004, 14, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Tsiftsoglou, S.A.; Arnold, J.N.; Roversi, P.; Crispin, M.D.; Radcliffe, C.; Lea, S.M.; Dwek, R.A.; Rudd, P.M.; Sim, R.B. Human complement factor I glycosylation: Structural and functional characterisation of the N-linked oligosaccharides. Biochim. Biophys. Acta 2006, 1764, 1757–1766. [Google Scholar] [CrossRef] [PubMed]
- Nelboeck, P.; Fuchs, M.; Bur, D.; Löffler, B.M. Glycosylation of Asn-632 and Asn-651 is important for functional expression of endothelin-converting enzyme-1. J. Cardiovasc. Pharmacol. 1998, 31, S4–S6. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, T.; Tsukuba, T.; Bertenshaw, G.P.; Bond, J.S. N-Linked Oligosaccharides on the Meprin A Metalloprotease Are Important for Secretion and Enzymatic Activity, but Not for Apical Targeting. J. Biol. Chem. 2000, 275, 25577–25584. [Google Scholar] [CrossRef] [PubMed]
- Gowda, D.C.; Jackson, C.M.; Kurzban, G.P.; McPhie, P.; Davidson, E.A. Core Sugar Residues of the N-Linked Oligosaccharides of Russell’s Viper Venom Factor X-Activator Maintain Functionally Active Polypeptide Structure. Biochemistry 1996, 35, 5833–5837. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Igarashi, T.; Mori, H. Crystal structure of RVV-X: An example of evolutionary gain of specificity by ADAM proteinases. FEBS Lett. 2007, 581, 5859–5864. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.I.; Munshi, H.G.; Sen, R.; Snipas, S.J.; Salvesen, G.S.; Fridman, R.; Stack, M.S. Glycosylation Broadens the Substrate Profile of Membrane Type 1 Matrix Metalloproteinase. J. Biol. Chem. 2004, 279, 8278–8289. [Google Scholar] [CrossRef] [PubMed]
- Chemical Computing Group. Molecular Operating Environment (MOE); 2015.10; Chemical Computing Group, Inc.: Montreal, QC, Canada, 2016. [Google Scholar]
- Barman, A.; Schürer, S.; Prabhakar, R. Computational Modeling of Substrate Specificity and Catalysis of the β-Secretase (BACE1) Enzyme. Biochemistry 2011, 50, 4337–4349. [Google Scholar] [CrossRef] [PubMed]
- Pelmenschikov, V.; Siegbahn, P.E.M. Catalytic Mechanism of Matrix Metalloproteinases: Two-Layered ONIOM Study. Inorg. Chem. 2002, 41, 5659–5666. [Google Scholar] [CrossRef] [PubMed]
- Hartley, B.S.; Neurath, H. Homologies in Serine Proteinases [and Discussion]. Philos. Trans. R. Soc. B 1970, 257, 77–87. [Google Scholar] [CrossRef]
- Ishida, T.; Kato, S. Theoretical Perspectives on the Reaction Mechanism of Serine Proteases: The Reaction Free Energy Profiles of the Acylation Process. J. Am. Chem. Soc. 2003, 125, 12035–12048. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Devi-Kesavan, L.S.; Gao, J. Molecular Dynamics Simulations of the Catalytic Pathway of a Cysteine Protease: A Combined QM/MM Study of Human Cathepsin K. J. Am. Chem. Soc. 2007, 129, 13633–13645. [Google Scholar] [CrossRef] [PubMed]
- Huber, E.M.; Heinemeyer, W.; Li, X.; Arendt, C.S.; Hochstrasser, M.; Groll, M. A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome. Nat. Commun. 2016, 7, 10900. [Google Scholar] [CrossRef] [PubMed]
- Chavaroche, A.; Cudic, M.; Giulianotti, M.; Houghten, R.A.; Fields, G.B.; Minond, D. Glycosylation of a disintegrin and metalloprotease 17 affects its activity and inhibition. Anal. Biochem. 2014, 449, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Minond, D.; Cudic, M.; Bionda, N.; Giulianotti, M.; Maida, L.; Houghten, R.A.; Fields, G.B. Discovery of Novel Inhibitors of a Disintegrin and Metalloprotease 17 (ADAM17) Using Glycosylated and Non-glycosylated Substrates. J. Biol. Chem. 2012, 287, 36473–36487. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yu, C.L.; Williams, S.R.; Springman, E.; Jeffery, D.; Sprengeler, P.A.; Estevez, A.; Sampang, J.; Shrader, W.; Spencer, J.; et al. Expression, Crystallization, and Three-dimensional Structure of the Catalytic Domain of Human Plasma Kallikrein. J. Biol. Chem. 2005, 280, 41077–41089. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, J.; Kalyan, N.K.; Lee, S.G.; Hum, W.T.; Rappaport, R.; Hung, P.P. Deglycosylation increases the fibrinolytic activity of a deletion mutant of tissue-type plasminogen activator. Thromb. Haemost. 1990, 63, 464–471. [Google Scholar] [PubMed]
- Howard, S.C.; Wittwer, A.J.; Welply, J.K. Oligosaccharides at each glycosylation site make structure-dependent contributions to biological properties of human tissue plasminogen activator. Glycobiology 1991, 1, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Wittwer, A.J.; Howard, S.C.; Carr, L.S.; Harakas, N.K.; Feder, J.; Parekh, R.B.; Rudd, P.M.; Dwek, R.A.; Rademacher, T.W. Effects of N-glycosylation on in vitro activity of Bowes melanoma and human colon fibroblast-derived tissue plasminogen activator. Biochemistry 1989, 28, 7662–7669. [Google Scholar] [CrossRef] [PubMed]
- Høiberg-Nielsen, R.; Westh, P.; Arleth, L. The Effect of Glycosylation on Interparticle Interactions and Dimensions of Native and Denatured Phytase. Biophys. J. 2009, 96, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Abouakil, N.; Mas, E.; Bruneau, N.; Benajiba, A.; Lombardo, D. Bile salt-dependent lipase biosynthesis in rat pancreatic AR 4–2 J cells. Essential requirement of N-linked oligosaccharide for secretion and expression of a fully active enzyme. J. Biol. Chem. 1993, 268, 25755–25763. [Google Scholar] [PubMed]
- Miller, G.C.; Long, C.J.; Bojilova, E.D.; Marchadier, D.; Badellino, K.O.; Blanchard, N.; Fuki, I.V.; Glick, J.M.; Rader, D.J. Role of N-linked glycosylation in the secretion and activity of endothelial lipase. J. Lipid Res. 2004, 45, 2080–2087. [Google Scholar] [CrossRef] [PubMed]
- Terzyan, S.; Wang, C.-S.; Downs, D.; Hunter, B.; Zhang, X.C. Crystal structure of the catalytic domain of human bile salt activated lipase. Protein Sci. 2000, 9, 1783–1790. [Google Scholar] [CrossRef] [PubMed]
- Wölle, J.; Jansen, H.; Smith, L.C.; Chan, L. Functional role of N-linked glycosylation in human hepatic lipase: Asparagine-56 is important for both enzyme activity and secretion. J. Lipid Res. 1993, 34, 2169–2176. [Google Scholar] [PubMed]
- Baldwin, E.T.; Bhat, T.N.; Gulnik, S.; Hosur, M.V.; Sowder, R.C.; Cachau, R.E.; Collins, J.; Silva, A.M.; Erickson, J.W. Crystal structures of native and inhibited forms of human cathepsin D: Implications for lysosomal targeting and drug design. Proc. Natl. Acad. Sci. USA 1993, 90, 6796–6800. [Google Scholar] [CrossRef] [PubMed]
- Turk, V.; Stoka, V.; Vasiljeva, O.; Renko, M.; Sun, T.; Turk, B.; Turk, D. Cysteine cathepsins: From structure, function and regulation to new frontiers. Biochim. Biophys. Acta 2012, 1824, 68–88. [Google Scholar] [CrossRef] [PubMed]
- Keil, C.; Maskos, K.; Than, M.; Hoopes, J.T.; Huber, R.; Tan, F.; Deddish, P.A.; Erdös, E.G.; Skidgel, R.A.; Bode, W. Crystal Structure of the Human Carboxypeptidase N (Kininase I) Catalytic Domain. J. Mol. Biol. 2007, 366, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Iino, M.; Foster, D.C.; Kisiel, W. Functional Consequences of Mutations in Ser-52 and Ser-60 in Human Blood Coagulation Factor VII. Arch. Biochem. Biophys. 1998, 352, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Sutkeviciute, I.; Mistiniene, E.; Sereikaite, J.; Bumelis, V.A. The influence of different glycosylation patterns on factor VII biological activity. Biochimie 2009, 91, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Böhm, E.; Seyfried, B.K.; Dockal, M.; Graninger, M.; Hasslacher, M.; Neurath, M.; Konetschny, C.; Matthiessen, P.; Mitterer, A.; Scheiflinger, F. Differences in N-glycosylation of recombinant human coagulation factor VII derived from BHK, CHO, and HEK293 cells. BMC Biotechnol. 2015, 15, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ries, M.; Easton, R.L.; Longstaff, C.; Zenker, M.; Morris, H.R.; Dell, A.; Gaffney, P.J. Differences between neonates and adults in carbohydrate sequences and reaction kinetics of plasmin and α2-antiplasmin. Thromb. Res. 2002, 105, 247–256. [Google Scholar] [CrossRef]
- Xue, Y.; Bodin, C.; Olsson, K. Crystal structure of the native plasminogen reveals an activation-resistant compact conformation. J. Thromb. Haemost. 2012, 10, 1385–1396. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Skala, W.; Magdolen, V.; Briza, P.; Biniossek, M.L.; Schilling, O.; Kellermann, J.; Brandstetter, H.; Goettig, P. A Single Glycan at the 99-Loop of Human Kallikrein-related Peptidase 2 Regulates Activation and Enzymatic Activity. J. Biol. Chem. 2016, 291, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Loke, I.; Packer, N.H.; Thaysen-Andersen, M. Complementary LC-MS/MS-Based N-Glycan, N-Glycopeptide, and Intact N-Glycoprotein Profiling Reveals Unconventional Asn71-Glycosylation of Human Neutrophil Cathepsin G. Biomolecules 2015, 5, 1832–1854. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, H.B.; Branner, S.; Wiberg, F.C.; Wagtmann, N. Crystal structure of human dipeptidyl peptidase IV/CD26 in complex with a substrate analog. Nat. Struct. Mol. Biol. 2003, 10, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.; Schomburg, I.; Placzek, S.; Jeske, L.; Ulbrich, M.; Xiao, M.; Sensen, C.W.; Schomburg, D. BRENDA in 2015: Exciting developments in its 25th year of existence. Nucleic Acids Res. 2015, 43, D439–D446. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Kato, S. Role of Asp102 in the Catalytic Relay System of Serine Proteases: A Theoretical Study. J. Am. Chem. Soc. 2004, 126, 7111–7118. [Google Scholar] [CrossRef] [PubMed]
- Toney, M.D. Common Enzymological Experiments Allow Free Energy Profile Determination. Biochemistry 2013, 52, 5952–5965. [Google Scholar] [CrossRef] [PubMed]
- Chu, W.-T.; Wang, J. Energy Landscape Topography Reveals the Underlying Link Between Binding Specificity and Activity of Enzymes. Sci. Rep. 2016, 6, 27808. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekhar, S. Thermodynamic analysis of enzyme catalysed reactions: New insights into the Michaelis–Menten equation. Res. Chem. Intermed. 2002, 28, 265–275. [Google Scholar] [CrossRef]
- De Castro, I.N.; Alonso, F.J. Energy diagrams for enzyme catalysed reactions: A confusing point in the textbooks. Biochem. Educ. 1997, 25, 87–89. [Google Scholar] [CrossRef]
- Hudáky, P.; Perczel, A. A self-stabilized model of the chymotrypsin catalytic pocket. The energy profile of the overall catalytic cycle. Proteins Struct. Funct. Bioinform. 2006, 62, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Scheiner, S.; Lipscomb, W.N. Molecular orbital studies of enzyme activity: Catalytic mechanism of serine proteinases. Proc. Natl. Acad. Sci. USA 1976, 73, 432–436. [Google Scholar] [CrossRef] [PubMed]
- Fersht, A.R.; Shi, J.-P.; Knill-Jones, J.; Lowe, D.M.; Wilkinson, A.J.; Blow, D.M.; Brick, P.; Carter, P.; Waye, M.M.Y.; Winter, G. Hydrogen bonding and biological specificity analysed by protein engineering. Nature 1985, 314, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.S.; Bokhari, S.A.; Afzal, A.J.; Singh, S. A Novel Thermodynamic Relationship Based on Kramers Theory for Studying Enzyme Kinetics under High Viscosity. IUBMB Life 2004, 56, 403–407. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, S.E.; Imperiali, B. A molecular basis for glycosylation-induced conformational switching. Chem. Biol. 1998, 5, 427–437. [Google Scholar] [CrossRef]
- Bosques, C.J.; Tschampel, S.M.; Woods, R.J.; Imperiali, B. Effects of Glycosylation on Peptide Conformation: A Synergistic Experimental and Computational Study. J. Am. Chem. Soc. 2004, 126, 8421–8425. [Google Scholar] [CrossRef] [PubMed]
- Skala, W.; Utzschneider, D.T.; Magdolen, V.; Debela, M.; Guo, S.; Craik, C.S.; Brandstetter, H.; Goettig, P. Structure-Function Analyses of Human Kallikrein-Related Peptidase 2 Establish the 99-Loop as Master Regulator of Activity. J. Biol. Chem. 2014, 289, 34267–34283. [Google Scholar] [CrossRef] [PubMed]
- Laxmikanthan, G.; Blaber, S.I.; Bernett, M.J.; Scarisbrick, I.A.; Juliano, M.A.; Blaber, M. 1.70 angstrom X-ray structure of human apo kallikrein 1: Structural changes upon peptide inhibitor/substrate binding. Proteins Struct. Funct. Bioinform. 2005, 58, 802–814. [Google Scholar] [CrossRef] [PubMed]
- Menez, R.; Michel, S.; Muller, B.H.; Bossus, M.; Ducancel, F.; Jolivet-Reynaud, C.; Stura, E.A. Crystal Structure of a Ternary Complex between Human Prostate-specific Antigen, Its Substrate Acyl Intermediate and an Activating Antibody. J. Mol. Biol. 2008, 376, 1021–1033. [Google Scholar] [CrossRef] [PubMed]
- Yoshimasu, M.A.; Tanaka, T.; Ahn, J.-K.; Yada, R.Y. Effect of N-linked glycosylation on the aspartic proteinase porcine pepsin expressed from Pichia pastoris. Glycobiology 2004, 14, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Shen, B.; Zhu, Z.; Zhang, P.; Ji, Y.; Niu, L.; Li, X.; Teng, M. Crystal structure and activating effect on RyRs of AhV_TL-I, a glycosylated thrombin-like enzyme from Agkistrodon halys snake venom. Arch. Toxicol. 2013, 87, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Vogt, A.D.; Di Cera, E. Conformational Selection Is a Dominant Mechanism of Ligand Binding. Biochemistry 2013, 52, 5723–5729. [Google Scholar] [CrossRef] [PubMed]
- Hammes, G.G.; Chang, Y.-C.; Oas, T.G. Conformational selection or induced fit: A flux description of reaction mechanism. Proc. Natl. Acad. Sci. USA 2009, 106, 13737–13741. [Google Scholar] [CrossRef] [PubMed]
- Michel, D. Conformational selection or induced fit? New insights from old principles. Biochimie 2016, 128–129, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Pol-Fachin, L.; Verli, H. Structural glycobiology of heparin dynamics on the exosite 2 of coagulation cascade proteases: Implications for glycosaminoglycans antithrombotic activity. Glycobiology 2014, 24, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Halabi, N.; Rivoire, O.; Leibler, S.; Ranganathan, R. Protein Sectors: Evolutionary Units of Three-Dimensional Structure. Cell 2009, 138, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Marti, T.; Schaller, J.; Rickli, E.E.; Schmid, K.; Kamerling, J.P.; Gerwig, G.J.; van Halbeek, H.; Vliegenthart, J.F.G. The N-and O-linked carbohydrate chains of human, bovine and porcine plasminogen. Eur. J. Biochem. 1988, 173, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Davidson, D.J.; Castellino, F.J. The influence of the nature of the asparagine 289-linked oligosaccharide on the activation by urokinase and lysine binding properties of natural and recombinant human plasminogens. J. Clin. Investig. 1993, 92, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Rudd, P.M.; Woods, R.J.; Wormald, M.R.; Opdenakker, G.; Downing, A.K.; Campbell, I.D.; Dwek, R.A. The effects of variable glycosylation on the functional activities of ribonuclease, plasminogen and tissue plasminogen activator. Biochim. Biophys. Acta 1995, 1248, 1–10. [Google Scholar] [CrossRef]
- Parekh, R.B.; Dwek, R.A.; Rudd, P.M.; Thomas, J.R.; Rademacher, T.W.; Warren, T.; Wun, T.C.; Hebert, B.; Reitz, B. N-Glycosylation and in vitro enzymic activity of human recombinant tissue plasminogen activator expressed in Chinese hamster ovary cells and a murine cell line. Biochemistry 1989, 28, 7670–7679. [Google Scholar] [CrossRef] [PubMed]
- Asselbergs, F.A.M.; Bürgi, R.; van Oostrum, J. Functional effects of kringle 2 glycosylation in a hybrid plasminogen activator. Blood Coagul. Fibrinolysis 1993, 4, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Bowman, K.K.; Clark, J.; Yu, L.; Mortara, K.; Radika, K.; Wang, J.; Zhan, H. Expression, Purification, and Characterization of Deglycosylated Human Pro-Prostate-Specific Antigen. Protein Expr. Purif. 2000, 20, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.-M.; Leinonen, J.; Kalkkinen, N.; Stenman, U.-H. Prostate-specific antigen forms a complex with and cleaves α1-protease inhibitor in vitro. Prostate 1997, 33, 87–96. [Google Scholar] [CrossRef]
- Gilgunn, S.; Conroy, P.J.; Saldova, R.; Rudd, P.M.; O’Kennedy, R.J. Aberrant PSA glycosylation—A sweet predictor of prostate cancer. Nat. Rev. Urol. 2013, 10, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Kuzmanov, U.; Jiang, N.X.; Smith, C.R.; Soosaipillai, A.; Diamandis, E.P. Differential N-glycosylation of Kallikrein 6 Derived from Ovarian Cancer Cells or the Central Nervous System. Mol. Cell. Proteom. 2009, 8, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Kuzmanov, U.; Smith, C.R.; Batruch, I.; Soosaipillai, A.; Diamandis, A.; Diamandis, E.P. Separation of kallikrein 6 glycoprotein subpopulations in biological fluids by anion-exchange chromatography coupled to ELISA and identification by mass spectrometry. Proteomics 2012, 12, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Behnken, H.N.; Ruthenbeck, A.; Schulz, J.-M.; Meyer, B. Glycan Analysis of Prostate Specific Antigen (PSA) Directly from the Intact Glycoprotein by HR-ESI/TOF-MS. J. Proteome Res. 2014, 13, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- White, K.Y.; Rodemich, L.; Nyalwidhe, J.O.; Comunale, M.A.; Clements, M.A.; Lance, R.S.; Schellhammer, P.F.; Mehta, A.S.; Semmes, O.J.; Drake, R.R. Glycomic Characterization of Prostate-Specific Antigen and Prostatic Acid Phosphatase in Prostate Cancer and Benign Disease Seminal Plasma Fluids. J. Proteome Res. 2009, 8, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Bhanushali, P.B.; Badgujar, S.B.; Tripathi, M.M.; Gupta, S.; Murthy, V.; Krishnasastry, M.V.; Puri, C.P. Development of glycan specific lectin based immunoassay for detection of prostate specific antigen. Int. J. Biol. Macromol. 2016, 86, 468–480. [Google Scholar] [CrossRef] [PubMed]
- Fuster, M.M.; Esko, J.D. The sweet and sour of cancer: Glycans as novel therapeutic targets. Nat. Rev. Cancer 2005, 5, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Anugraham, M.; Jacob, F.; Nixdorf, S.; Everest-Dass, A.V.; Heinzelmann-Schwarz, V.; Packer, N.H. Specific Glycosylation of Membrane Proteins in Epithelial Ovarian Cancer Cell Lines: Glycan Structures Reflect Gene Expression and DNA Methylation Status. Mol. Cell. Proteom. 2014, 13, 2213–2232. [Google Scholar] [CrossRef] [PubMed]
- Żurawska-Płaksej, E.; Kratz, E.M.; Ferens-Sieczkowska, M.; Knapik-Kordecka, M.; Piwowar, A. Changes in glycosylation of human blood plasma chitotriosidase in patients with type 2 diabetes. Glycoconj. J. 2016, 33, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, P.P.; Callewaert, N. N-glycosylation engineering of biopharmaceutical expression systems. Curr. Mol. Med. 2009, 9, 774–800. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.G.; Lloyd, R.C.; Jones, J.B. Controlled site-selective protein glycosylation for precise glycan structure–catalytic activity relationships. Bioorg. Med. Chem. 2000, 8, 1527–1535. [Google Scholar] [CrossRef]
- Stura, E.A.; Muller, B.H.; Bossus, M.; Michel, S.; Jolivet-Reynaud, C.; Ducancel, F. Crystal structure of human prostate-specific antigen in a sandwich antibody complex. J. Mol. Biol. 2011, 414, 530–544. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Walczak, M.J.; Aebi, M.; Wider, G. Posttranslational Modifications of Intact Proteins Detected by NMR Spectroscopy: Application to Glycosylation. Angew. Chem. Int. Ed. 2015, 54, 7096–7100. [Google Scholar] [CrossRef] [PubMed]






| Family | UniProtKB | Aspartic Proteases | PDB (N-/O-Glycans) |
|---|---|---|---|
| A1 | BACE1_HUMAN | β-Secretase 1, 2 | - |
| RENI_HUMAN | Renin | 1HRN (1N) | |
| CATD_HUMAN | Cathepsin D(o), E | D: 1LYA (2N), 1LYB (2N) | |
| NAPSA_HUMAN | Napsin-A | - | |
| A22 | SPP2A_HUMAN | Signal peptide peptidase-like 2A, 2B, 2C | - |
| HM13_HUMAN | Minor histocompatibility antigen H13 | - | |
| A28 | APRV1_HUMAN | Retroviral-like aspartic protease 1 | - |
| Cysteine Proteases | |||
| C1 | CATB_HUMAN | Cathepsin B, C, F, H, K, L2, S, W, Z | C: 1K3B (4N), 2DJF (4N) |
| C12 | UCHL1_HUMAN | Ubiquitin C-terminal hydrolase isozyme L1(o) | - |
| C13 | LGMN_HUMAN | Legumain | 4AW9 (3N) |
| C26 | GGH_HUMAN | γ-Glutamyl hydrolase | - |
| Family | UniProtKB | Metallo Proteases | PDB (N-/O-Glycans) |
|---|---|---|---|
| M1 | AMPE_HUMAN | Aminopeptidase E, N(o), Q | E: 4KX7 (8N), N: 4FYT (8N) |
| ERAP1_HUMAN | Endoplasmic reticulum aminopeptidase 1, 2 | 1: 2YD0 (3N), 2: 5AB0 (8N) | |
| LCAP_HUMAN | Leucyl-cystinyl aminopeptidase | 4P8Q (8N) | |
| TRHDE_HUMAN | TRH-degrading ectoenzyme | - | |
| M2 | ACE_HUMAN | Angiotensin-converting enzyme 1, ACE 2 | 1O8A (6N), ACE2: 2AJF (3N) |
| M10 | MMP1_HUMAN | MMP 1–3, 8, 9(o), 12, 13, 14(o), 15–17, 19, 21, 23, 26–28 | - |
| M12 | ADAM2_HUMAN | ADAM 2, 7–10, 12, 15, 17–23, 28–30, 32, 33 | 33: 1R55 (1N) |
| ADEC1_HUMAN | ADAM DEC1 | - | |
| ATS1_HUMAN | ADAM-TS 1–4, 5(o), 6-10, 12, 13(o), 14–20 | 5: 2RJQ (1N) 13: 3GHN (3N/1O) | |
| BMP1_HUMAN | Bone morphogenetic protein 1 | - | |
| TLL1_HUMAN | Tolloid-like protein 1, 2 | - | |
| MEP1A_HUMAN | Meprin A subunit α, β(o) | 4GWM (7N) | |
| M13 | ECE1_HUMAN | Endothelin-converting enzyme 1, 2, -like 1 | - |
| NEP_HUMAN | Neprilysin | 5JMY (4N) | |
| KELL_HUMAN | Kell protein | - | |
| MMEL1_HUMAN | Membrane metallo-endopeptidase-like 1 | - | |
| PHEX_HUMAN | Phosphate-regulating neutral endopeptidase | - | |
| M14 | CBPA4_HUMAN | Carboxypeptidase A4, A6, B2, D, E | A4: 2BOA (1N),B2: 3D68 (4N) |
| Carboxypeptidase M, N(o), O, X1, Z | M: 1UWY (1N) , N: 2NSM (3O) | ||
| M16 | IDE_HUMAN | Insulin-degrading enzyme | - |
| M19 | DPEP1_HUMAN | Dipeptidase 1, 2, 3 | 1: 1ITQ (2N) |
| M20 | P20D1_HUMAN | Carboxypeptidase PM20D1 | - |
| CNDP1_HUMAN | β-Ala-His dipeptidase | - | |
| M24 | MAP2_HUMAN | Methionine aminopeptidase 2(o) | - |
| XPP2_HUMAN | Xaa-Pro aminopeptidase 2 | - | |
| M28 | CBPQ_HUMAN | Carboxypeptidase Q | - |
| ERMP1_HUMAN | Endoplasmic reticulum metallopeptidase 1 | - | |
| FOLH1_HUMAN | Glutamate carboxypeptidase 2 | 2C6C (6N) | |
| NALD2_HUMAN | NAALADase 2, NAALADaseL | 2: 3FED (4N), L: 4TWE (6N) | |
| M43 | PAPP1_HUMAN | Pappalysin 1, 2 | - |
| M50 | MBTP1_HUMAN | MB transcription factor site protease 1, 2 | - |
| M87 | CLCA1_HUMAN | Ca-activated chloride channel regulator 1, 2, 4 | - |
| M96 | TIKI1_HUMAN | Metalloprotease TIKI1, TIKI2 | - |
| Family | UniProtKB | Serine Proteases | PDB (N-/O-Glycans) |
|---|---|---|---|
| S1 | C1R_HUMAN | Complement C1r, C1s, C2 C1r subcomponent-like protein | 1r: 1GPZ (2N), 1s: 1ELV (1N) 2: 2I6S (5N) |
| CFAB_HUMAN | Complement factor B, I | B: 2OK5 (4N), I: 2XRC (6N) | |
| CELA1_HUMAN | Chymotrypsin-like elastase family A1, 3A, 3B | - | |
| CMA1_HUMAN | Chymase | 1NN6 (2N) | |
| CORIN_HUMAN | Atrial natriuretic peptide-converting enzyme | - | |
| CTRC_HUMAN | Chymotrypsin-C, CTR-like protease 1 | - | |
| ELNE_HUMAN | Neutrophil elastase | 1PPG (2N) | |
| ENTK_HUMAN | Enteropeptidase (Enterokinase) | ||
| FA7_HUMAN | Coagulation factor V, VII(o), IX(o), X(o) | 7: 1QFK (1N/2O) 9: 3KCG (3N) | |
| Coagulation factor XI, XII(o) | 11: 5EOK (4N) 12: 4XE4 (1N) | ||
| GRAA_HUMAN | Granzyme A, B, H, M | B: 1IAU (2N) | |
| HABP2_HUMAN | Hyaluronan-binding protein 2 | - | |
| HEPS_HUMAN | Serine protease hepsin | - | |
| HGFA_HUMAN | Hepatocyte growth factor activator | 2R0L (1N) | |
| KLK1_HUMAN | Kallikrein-related peptidase 1(o), 2, 3(o), 4, 5 | 1: 1SPJ (1N) 3: 3QUM (1N/1O) | |
| 6–13, 15 | 5: 2PSX (1N) | ||
| KLKB1_HUMAN | Plasma kallikrein | - | |
| MASP1_HUMAN | Mannan-binding lectin serine protease 1 | 3DEM (1N) | |
| NETR_HUMAN | Neurotrypsin | - | |
| OVCH1_HUMAN | Ovochymase-1, 2 | - | |
| PLMN_HUMAN | Plasminogen(o) | 4DUR (1O) | |
| POLS2_HUMAN | Polyserase-2 | - | |
| PROC_HUMAN | Vitamin K-dependent protein C(o) | - | |
| PRS23_HUMAN | SP 23, 27, 29, 35, 38, 41, 42, 44, 45, 47, 48, 55–58 | 57: 4Q7Y (2N) | |
| PRSS8_HUMAN | Prostasin | - | |
| PRTN3_HUMAN | Myeloblastin | 1FUJ (4N) | |
| ST14_HUMAN | Matriptase | - | |
| TEST_HUMAN | Testisin | - | |
| THRB_HUMAN | Prothrombin | 5E8E (1N) | |
| TMPS2_HUMAN | Transmembrane (TM) protease serine 2–7, 9 | - | |
| TMPSC_HUMAN | TM protease serine 12, 13 | - | |
| TM11A_HUMAN | TM protease serine 11A, 11B, 11D, 11E | - | |
| TPA_HUMAN | Tissue-type plasminogen activator(o) | - | |
| TRYB1_HUMAN | Tryptase α/β-1, β-2, γ, δ | B1: 2F9N (2N) | |
| UROK_HUMAN | Urokinase-type plasminogen activator(o) | 2FD6 (3N) |
| Family | UniProtKB | Serine Proteases | PDB (N-/O-Glycans) |
|---|---|---|---|
| S8 | FURIN_HUMAN | Furin | - |
| NEC1_HUMAN | Neuroendocrine convertase 1(o), 2 | - | |
| PCSK4_HUMAN | PPC subtilisin/kexin type 4–7, 9 | 9: 4NE9 (1N) | |
| S9 | DPP4_HUMAN | Dipeptidyl peptidase 4 | 1N1M (6) |
| SEPR_HUMAN | Prolyl endopeptidase FAP | 1Z68 (5N) | |
| S10 | PPGB_HUMAN | Cathepsin A | 4MWS (3N) |
| RISC_HUMAN | Retinoid-inducible serine carboxypeptidase | - | |
| S28 | DPP2_HUMAN | Dipeptidyl peptidase | 3JYH (4N) |
| PCP_HUMAN | Lysosomal Pro-X carboxypeptidase | 3N2Z (5N) | |
| TSSP_HUMAN | Thymus-specific serine protease | - | |
| S53 | TPP1_HUMAN | Tripeptidyl-peptidase 1 | 3EE6 (4N) |
| S60 | TRFL_HUMAN | Lactotransferrin | 1LGB (1N) |
| Threonine Proteases | |||
| T1 | PSA1_HUMAN | Proteasome subunit α type-1(o), 5–7(o) | - |
| PSB1_HUMAN | Proteasome subunit β type-1(o), 6(o) | - | |
| T3 | GGT1_HUMAN | γ-Glutamyltransferase 1, 3, 5–7 | 1: 4GDX (6N) |
© 2016 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goettig, P. Effects of Glycosylation on the Enzymatic Activity and Mechanisms of Proteases. Int. J. Mol. Sci. 2016, 17, 1969. https://doi.org/10.3390/ijms17121969
Goettig P. Effects of Glycosylation on the Enzymatic Activity and Mechanisms of Proteases. International Journal of Molecular Sciences. 2016; 17(12):1969. https://doi.org/10.3390/ijms17121969
Chicago/Turabian StyleGoettig, Peter. 2016. "Effects of Glycosylation on the Enzymatic Activity and Mechanisms of Proteases" International Journal of Molecular Sciences 17, no. 12: 1969. https://doi.org/10.3390/ijms17121969
APA StyleGoettig, P. (2016). Effects of Glycosylation on the Enzymatic Activity and Mechanisms of Proteases. International Journal of Molecular Sciences, 17(12), 1969. https://doi.org/10.3390/ijms17121969