Knockdown of ELMO3 Suppresses Growth, Invasion and Metastasis of Colorectal Cancer
Abstract
:1. Introduction
2. Results
2.1. Expression of ELMO3 Was Aberrant in Human Colorectal Cancer (CRC) Tissues
2.2. The Correlation between ELMO3 Expression and Clinicopathological Characteristics
2.3. Knockdown of ELMO3 Inhibits Cell Proliferation of HCT116 Cells
2.4. Knockdown of ELMO3 Induces G1 Cell Cycle Arrest
2.5. Knockdown of ELMO3 Supresses Cell Invasion and Metastasis
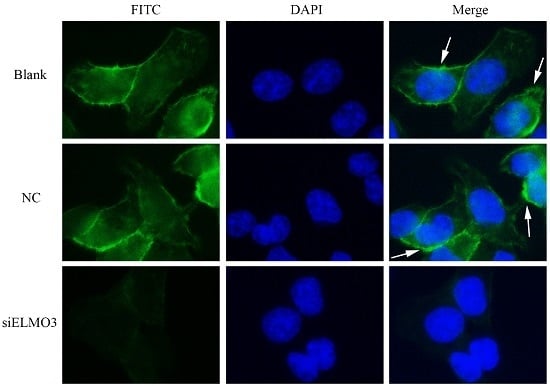
2.6. Knockdown ELMO3 Supresses the F-Actin Polymerization of HCT116 Cells
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.2. RT-PCR and qPCR Analysis
4.3. Western Blot Analysis
4.4. Immunohistochemical Analysis
4.5. Cell Culture and ELMO3 siRNA Transfection
4.6. MTS Assay
4.7. Cell Cycle Analysis
4.8. Wound Healing Assays
4.9. Transwell Migration and Invasion Assay
4.10. Immunofluorescence Staining
4.11. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Anitha, A.; Maya, S.; Sivaram, A.J.; Mony, U.; Jayakumar, R. Combinatorial nanomedicines for colon cancer therapy. Wires Nanomed. Nanobiotechnol. 2016, 8, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, D.; Atkin, W.; Lenz, H.-J.; Lynch, H.T.; Minsky, B.; Nordlinger, B.; Starling, N. Colorectal cancer. Lancet 2010, 375, 1030–1047. [Google Scholar] [CrossRef]
- Lin, C.C.; Ng, H.L.H.; Pan, W.S.; Chen, H.B.; Zhang, G.; Bian, Z.X.; Lu, A.P.; Yang, Z.J. Exploring different strategies for efficient delivery of colorectal cancer therapy. Int. J. Mol. Sci. 2015, 16, 26936–26952. [Google Scholar] [CrossRef] [PubMed]
- Brody, H. Colorectal cancer. Nature 2015, 521, S1–S38. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Kanas, G.P.; Taylor, A.; Primrose, J.N.; Langeberg, W.J.; Kelsh, M.A.; Mowat, F.S.; Alexander, D.D.; Choti, M.A.; Poston, G. Survival after liver resection in metastatic colorectal cancer: Review and meta-analysis of prognostic factors. Clin. Epidemiol. 2012, 4, 283–301. [Google Scholar] [PubMed]
- Nicolson, G.L. Paracrine and autocrine growth mechanisms in tumor metastasis to specific sites with particular emphasis on brain and lung metastasis. Cancer Metastasis Rev. 1993, 12, 325–343. [Google Scholar] [CrossRef] [PubMed]
- Zlotnik, A. Involvement of chemokine receptors in organ-specific metastasis. Contrib. Microbiol. 2006, 13, 191–199. [Google Scholar] [PubMed]
- Iijima, M.; Devreotes, P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell 2002, 109, 599–610. [Google Scholar] [CrossRef]
- Yan, J.; Mihaylov, V.; Xu, X.; Brzostowski, J.A.; Li, H.; Liu, L.; Veenstra, T.D.; Parent, C.A.; Jin, T. A Gβγ effector, ELMOE, transduces GPCR signaling to the actin network during chemotaxis. Dev. Cell 2012, 22, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Gumienny, T.L.; Brugnera, E.; Tosello-Trampont, A.C.; Kinchen, J.M.; Haney, L.B.; Nishiwaki, K.; Walk, S.F.; Nemergut, M.E.; Macara, I.G.; Francis, R.; et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 2001, 107, 27–41. [Google Scholar] [CrossRef]
- Geisbrecht, E.R.; Haralalka, S.; Swanson, S.K.; Florens, L.; Washburn, M.P.; Abmayr, S.M. Drosophila ELMO/CED-12 interacts with Myoblast city to direct myoblast fusion and ommatidial organization. Dev. Biol. 2008, 314, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Brzostowski, J.A.; Fey, P.; Yan, J.; Isik, N.; Jin, T. The ELMO family forms an ancient group of actin-regulating proteins. Commun. Integr. Biol. 2009, 2, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Pelletier, A.; Cote, J.F. Opening up on ELMO regulation: New insights into the control of Rac signaling by the DOCK180/ELMO complex. Small GTPases 2011, 2, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Margaron, Y.; Fradet, N.; Yang, Q.; Wilkes, B.; Bouvier, M.; Hofmann, K.; Cote, J.F. An evolutionarily conserved autoinhibitory molecular switch in ELMO proteins regulates Rac signaling. Curr. Biol. 2010, 20, 2021–2027. [Google Scholar] [CrossRef] [PubMed]
- Jarzynka, N.J.; Hu, B.; Hui, K.M.; Bar-Joseph, I.; Gu, W.S.; Hirose, T.; Haney, L.B.; Ravichandran, K.S.; Nishikawa, R.; Cheng, S.Y. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Res. 2007, 67, 7203–7211. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Shi, L.; Lu, S.; Sun, X.; Liu, Y.; Li, H.; Wang, X.; Zhao, C.; Zhang, H.; Wang, Y. Autocrine IL-8 promotes F-actin polymerization and mediate mesenchymal transition via ELMO1-NF-κB-Snail signaling in glioma. Cancer Biol. Ther. 2015, 16, 898–911. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, L.; Fu, H.; Yan, J.; Wang, Y.; Guo, H.; Hao, X.; Xu, X.; Jin, T.; Zhang, N. Association between Galphai2 and ELMO1/Dock180 connects chemokine signalling with Rac activation and metastasis. Nat. Commun. 2013, 4, 1706. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.R.; Liu, G.Q.; Miao, X.Y.; Hua, S.W.; Zhong, D.W. Overexpression of engulfment and cell motility 1 promotes cell invasion and migration of hepatocellular carcinoma. Exp. Ther. Med. 2011, 2, 505–511. [Google Scholar] [PubMed]
- Wang, J.; Dai, J.M.; Che, Y.L.; Gao, Y.M.; Peng, H.J.; Liu, B.; Wang, H.; Hua, L.H. ELMO1 Helps Dock180 to Regulate Rac1 Activity and Cell Migration of Ovarian Cancer. Int. J. Gynecol. Cancer 2014, 24, 844–850. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Linghu, H.; Wang, J.; Che, Y.L.; Xiang, T.X.; Tang, W.X.; Yao, Z.W. The role of Crk/Dock180/Rac1 pathway in the malignant behavior of human ovarian cancer cell SKOV3. Tumor Biol. 2010, 31, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Dulak, A.M.; Stojanov, P.; Peng, S.Y.; Lawrence, M.S.; Fox, C.; Stewart, C.; Bandla, S.; Imamura, Y.; Schumacher, S.E.; Shefler, E.; et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat. Genet. 2013, 45, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Rapa, E.; Hill, S.K.; Morten, K.J.; Potter, M.; Mitchell, C. The over-expression of cell migratory genes in alveolar rhabdomyosarcoma could contribute to metastatic spread. Clin. Exp. Metastasis 2012, 29, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, C.; de la Rosa, G.; Anderson, C.S.; Murphy, P.S.; Capece, T.; Kim, M.; Elliott, M.R. Essential role of ELMO1 in Dock2-dependent lymphocyte migration. J. Immunol. 2014, 192, 6062–6070. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, X.; Pan, M.; Jin, T. ELMO1 Directly Interacts with Gβγ Subunit to Transduce GPCR Signaling to Rac1 Activation in Chemotaxis. J. Cancer 2016, 7, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Epting, D.; Slanchev, K.; Boehlke, C.; Hoff, S.; Loges, N.T.; Yasunaga, T.; Indorf, L.; Nestel, S.; Lienkamp, S.S.; Omran, H.; et al. The Rac1 regulator ELMO controls basal body migration and docking in multiciliated cells through interaction with Ezrin. Development 2015, 142, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Franke, K.; Otto, W.; Johannes, S.; Baumgart, J.; Nitsch, R.; Schumacher, S. miR-124-regulated RhoG reduces neuronal process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. EMBO J. 2012, 31, 2908–2921. [Google Scholar] [CrossRef] [PubMed]
- East, M.P.; Bowzard, J.B.; Dacks, J.B.; Kahn, R.A. ELMO Domains, Evolutionary and Functional Characterization of a Novel GTPase-activating Protein (GAP) Domain for ARF Protein Family GTPases. J. Biol. Chem. 2012, 287, 39538–39553. [Google Scholar] [CrossRef] [PubMed]
- Grimsley, C.M.; Kinchen, J.M.; Tosello-Trampont, A.C.; Brugnera, E.; Haney, L.B.; Lu, M.; Chen, Q.; Klingele, D.; Hengartner, M.O.; Ravichandran, K.S. Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration. J. Biol. Chem. 2004, 279, 6087–6097. [Google Scholar] [CrossRef] [PubMed]
- Toret, C.P.; Collins, C.; Nelson, W.J. An ELMO-Dock complex locally controls Rho GTPases and actin remodeling during cadherin-mediated adhesion. J. Cell Biol. 2014, 207, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Yang, L. The Functions of ELMO2 on Hepatocellular Carcinoma Cells Migration and Invasion. Master’s Thesis, Tianjin Medical University, Tianjin, China, 31 May 2013. [Google Scholar]
- Fan, W.; Yang, H.K.; Xue, H.; Sun, Y.; Zhang, J. ELMO3 is a novel biomarker for diagnosis and prognosis of non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 5503–5508. [Google Scholar] [PubMed]
- Soes, S.; Daugaard, I.L.; Sorensen, B.S.; Carus, A.; Mattheisen, M.; Alsner, J.; Overgaard, J.; Hager, H.; Hansen, L.L.; Kristensen, L.S. Hypomethylation and increased expression of the putative oncogene ELMO3 are associated with lung cancer development and metastases formation. Oncoscience 2014, 1, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Soes, S.; Hansen, L.L. ELMO3: A direct driver of cancer metastasis? Cell Cycle 2014, 13, 2483–2484. [Google Scholar] [CrossRef] [PubMed]
- Goyette, M.A.; Cote, J.F. NSCLC metastasis: Going with ELMO3. Oncotarget 2014, 5, 5850–5851. [Google Scholar] [CrossRef] [PubMed]
- Coskun, M.; Boyd, M.; Olsen, J.; Troelsen, J.T. Control of Intestinal Promoter Activity of the Cellular Migratory Regulator Gene ELMO3 by CDX2 and SP1. J. Cell. Biochem. 2010, 109, 1118–1128. [Google Scholar] [PubMed]
- Olson, M.F.; Sahai, E. The actin cytoskeleton in cancer cell motility. Clin. Exp. Metastasis 2009, 26, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Abu-Thuraia, A.; Gauthier, R.; Chidiac, R.; Fukui, Y.; Screaton, R.A.; Gratton, J.P.; Cote, J.F. Axl phosphorylates ELMO scaffold proteins to promote Rac activation and cell invasion. Mol. Cell. Biol. 2015, 35, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Duronio, R.J.; Xiong, Y. Signaling pathways that control cell proliferation. Cold Spring Harb. Perspect. Biol. 2013, 5, a008904. [Google Scholar] [CrossRef] [PubMed]
- Coskun, M.; Troelsen, J.T.; Nielsen, O.H. The role of CDX2 in intestinal homeostasis and inflammation. Biochim. Biophys. Acta 2011, 1812, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Laurin, M.; Huber, J.; Pelletier, A.; Houalla, T.; Park, M.; Fukui, Y.; Haibe-Kains, B.; Muller, W.J.; Cote, J.F. Rac-specific guanine nucleotide exchange factor DOCK1 is a critical regulator of HER2-mediated breast cancer metastasis. Proc. Natl. Acad. Sci. USA 2013, 110, 7434–7439. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Shen, W.; Zhou, H.; Dong, W.; Gao, D. Knockdown of LI-cadherin alters expression of matrix metalloproteinase-2 and -9 and galectin-3. Mol. Med. Rep. 2016, 13, 4469–4474. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Xu, Z.; Kuang, X.; Qiao, P.; Liu, S.; Zhang, L.; He, P.; Jadwiga, W.S.; Wang, Y.; Min, W. Molecular characterization and expression analysis of the autophagic gene Beclin 1 from the purse red common carp (Cyprinus carpio) exposed to cadmium. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2014, 160, 15–22. [Google Scholar] [CrossRef] [PubMed]






| Groups | N | ELMO3 Immunoreactivity Score | Positive Rate (%) | |||
|---|---|---|---|---|---|---|
| − | + | ++ | +++ | |||
| Tumor | 45 | 5 | 9 | 12 | 19 | 88.89% ** |
| Normal | 45 | 28 | 9 | 6 | 2 | 37.78% |
| Clinical Parameter | ELMO3 Immunoreactivity Score | N | p-Value | |||
|---|---|---|---|---|---|---|
| − | + | ++ | +++ | |||
| Gender | 0.883 † | |||||
| Male | 5 | 5 | 4 | 13 | 27 | |
| Female | 0 | 4 | 8 | 6 | 18 | |
| Age (years) | ||||||
| ≤60 | 3 | 6 | 9 | 7 | 25 | 0.094 † |
| >60 | 2 | 3 | 3 | 12 | 20 | |
| Tumor size (cm) | 0.007 † | |||||
| ≤4.0 | 4 | 8 | 7 | 7 | 26 | |
| >4.0 | 1 | 1 | 5 | 12 | 19 | |
| Differentiation | 0.001 ‡ | |||||
| High | 4 | 3 | 1 | 0 | 8 | |
| Moderate | 1 | 5 | 10 | 12 | 28 | |
| Poor | 0 | 1 | 1 | 7 | 9 | |
| Depth of invasion | 0.009 † | |||||
| T1 + T2 | 4 | 1 | 4 | 1 | 10 | |
| T3 + T4 | 1 | 8 | 8 | 18 | 35 | |
| Lymph node metastasis | 0.003 † | |||||
| No | 5 | 9 | 9 | 10 | 33 | |
| Yes | 0 | 0 | 3 | 9 | 12 | |
| Distant metastasis | 0.013 † | |||||
| No | 5 | 9 | 12 | 14 | 40 | |
| Yes | 0 | 0 | 0 | 5 | 5 | |
| TNM classification | 0.000 † | |||||
| I + II | 5 | 9 | 9 | 7 | ||
| III + IV | 0 | 0 | 3 | 11 | ||
| Lymph node positive number | 0.003 † | |||||
| <1 | 5 | 9 | 9 | 10 | 33 | |
| ≥1 | 0 | 0 | 3 | 9 | 12 | |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, H.-Y.; Yu, Q.-F.; Shen, W.; Guo, C.-M.; Li, Z.; Zhou, X.-Y.; Zhou, N.-J.; Min, W.-P.; Gao, D. Knockdown of ELMO3 Suppresses Growth, Invasion and Metastasis of Colorectal Cancer. Int. J. Mol. Sci. 2016, 17, 2119. https://doi.org/10.3390/ijms17122119
Peng H-Y, Yu Q-F, Shen W, Guo C-M, Li Z, Zhou X-Y, Zhou N-J, Min W-P, Gao D. Knockdown of ELMO3 Suppresses Growth, Invasion and Metastasis of Colorectal Cancer. International Journal of Molecular Sciences. 2016; 17(12):2119. https://doi.org/10.3390/ijms17122119
Chicago/Turabian StylePeng, Hui-Yun, Qiong-Fang Yu, Wei Shen, Cheng-Ming Guo, Zhen Li, Xiao-Yan Zhou, Nan-Jin Zhou, Wei-Ping Min, and Dian Gao. 2016. "Knockdown of ELMO3 Suppresses Growth, Invasion and Metastasis of Colorectal Cancer" International Journal of Molecular Sciences 17, no. 12: 2119. https://doi.org/10.3390/ijms17122119
APA StylePeng, H. -Y., Yu, Q. -F., Shen, W., Guo, C. -M., Li, Z., Zhou, X. -Y., Zhou, N. -J., Min, W. -P., & Gao, D. (2016). Knockdown of ELMO3 Suppresses Growth, Invasion and Metastasis of Colorectal Cancer. International Journal of Molecular Sciences, 17(12), 2119. https://doi.org/10.3390/ijms17122119