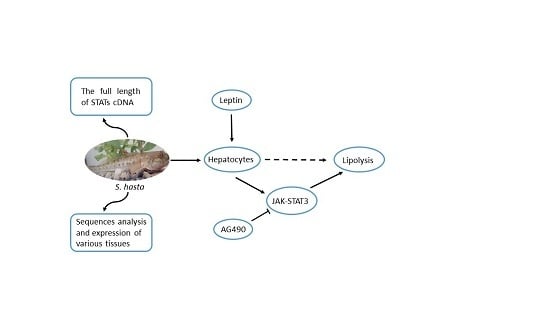
Isolation and Expression Analysis of STAT Members from Synechogobius hasta and Their Roles in Leptin Affecting Lipid Metabolism
Abstract
:1. Introduction
2. Results
2.1. Complete Sequences and Phylogenetic Analysis of Signal Transducers and Activators of Transcription (STATs)
2.2. STATs Structure and Sequence Analysis
2.3. Expression of S. hasta STATs in Various Tissues
2.4. Cell Viability and Intracellular Triglyceride (TG) Content
2.5. Enzymatic Activities
2.6. Transcriptional Regulation of the Genes by Leptin in Vitro
3. Discussion
4. Materials and Methods
4.1. STAT cDNAs Cloning and mRNA Expression Patterns of Various Tissues
4.1.1. Fish Rearing and Sampling
4.1.2. RNA Isolation and cDNAs Synthesis
4.1.3. Cloning and Sequencing of STATs
4.1.4.Sequence Analysis
4.2. Hepatocytes Treatment in Vitro
4.2.1. Hepatocytes Culture and Treatment
4.2.2. Cell Viability Assay, TG Content and Enzyme Activity Determination
4.3. Quantitative Polymerase Chain Reaction (PCR)
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 6PGD | 6-phosphogluconate dehydrogenase |
| CPT I | carnitine palmitoyl transferase I |
| FAS | fatty acid synthase |
| G6PD | glucose-6-phosphate dehydrogenase |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| HPRT | hypoxanthine-guanine phosphoribosyl transferase |
| JAK | janus kinase |
| LepR | leptin receptor |
| MS-222 | tricaine methanesulfonate |
| RPL7 | ribosomal protein L7 |
| STAT | signal transducer and activator of transcription |
| TBP | TATA-box-binding protein |
| TG | triglyceride |
| TUBA | htubulin alpha chain |
| UBCE | ubiquitin-conjugating enzyme |
| WGB | whole genome duplication |
References
- Horvath, C.M. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem. Sci. 2000, 25, 496–502. [Google Scholar] [CrossRef]
- Guo, C.J.; Zhang, Y.F.; Yang, L.S.; Yang, X.B.; Wu, Y.Y.; Liu, D.; Chen, W.J.; Weng, S.P.; Yu, X.Q.; He, J.G. The JAK and STAT family members of the mandarin fish Siniperca. chuatsi: Molecular cloning, tissues distribution and immunobiological activity. Fish Shellfish Immunol. 2009, 27, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Tan, X.Y.; Xu, Y.H.; Chen, Q.L.; Pan, Y.X. JAK and STAT members of yellow catfish Pelteobagrus fulvidraco and their roles in leptin affecting lipid metabolism. Gen. Comp. Endocrinol. 2016, 226, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Park, E.M.; Kang, J.H.; Seo, J.S.; Kim, G.D.; Chung, J.; Choi, T.J. Molecular cloning and expression analysis of the STAT1 gene from olive flounder, Paralichthys olivaceus. BMC Immunol. 2008, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Collet, B.; Ganne, G.; Bird, S.; Collins, C.M. Isolation and expression profile of a gene encoding for the signal transducer and activator of transcription STAT2 in Atlantic salmon (Salmo salar). Dev. Comp. Immunol. 2009, 33, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Sung, S.C.; Fan, T.J.; Chou, C.M.; Leu, J.H.; Hsu, Y.L.; Chen, S.T.; Huang, C.J. Genomic structure, expression and characterization of a STAT5 homologue from pufferfish (Tetraodon fluviatilis). Eur. J. Biochem. 2003, 270, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yang, C.G.; Sun, Z.Z.; Wang, X.L.; Chen, S.L. Signal transducer and activator of transcription 3 (STAT3) homologue in turbot (Scophthalmus maximus): Molecular characterization and expression analysis. Fish Shellfish Immunol. 2011, 30, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Wang, X.L.; Yang, C.G.; Chen, S.L. Molecular cloning, subcelluar location and expression profile of signal transducer and activator of transcription 2 (STAT2) from turbot, Scophthalmus maximus. Fish Shellfish Immunol. 2013, 35, 1200–1208. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.; Van de Peer, Y. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD). Bioessays 2005, 27, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Gorissen, M.; de Vrieze, E.; Flik, G.; Huising, M.O. STAT genes display differential evolutionary rates that correlate with their roles in the endocrine and immune system. J. Endocrinol. 2011, 209, 175–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, C.P.; Cao, X. Structure, function, and regulation of STAT proteins. Mol. Biosyst. 2006, 2, 536–550. [Google Scholar] [CrossRef] [PubMed]
- De Pedro, N.; Martinez-Alvarez, R.; Delgado, M.J. Acute and chronic leptin reduces food intake and body weight in goldfish (Carassius auratus). J. Endocrinol. 2006, 188, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Li, G.G.; Liang, X.F.; Xie, Q.; Li, G.; Yu, Y.; Lai, K. Gene structure, recombinant expression and functional characterization of grass carp leptin. Gen. Comp. Endocrinol. 2010, 166, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, A.J.; Conde-Sieira, M.; Lopez-Patino, M.A.; Miguez, J.M.; Soengas, J.L. In vitro leptin treatment of rainbow trout hypothalamus and hindbrain affects glucosensing and gene expression of neuropeptides involved in food intake regulation. Peptides 2011, 32, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.H.; Liang, X.F.; Wang, M.; Zhou, Y.; Bai, X.L.; He, Y. The role of leptin in lipid metabolism in fatty degenerated hepatocytes of the grass carp Ctenopharyngodon idellus. Fish Physiol. Biochem. 2012, 38, 1759–1774. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.F.; Wu, K.; Tan, X.Y.; Zhang, L.H.; Zhuo, M.Q.; Pan, Y.X.; Chen, Q.L. Effects of recombinant human leptin administration on hepatic lipid metabolism in yellow catfish Pelteobagrus fulvidraco: In vivo and in vitro studies. Gen. Comp. Endocrinol. 2015, 212, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Li, X.; Bai, H.; Gong, S. Effects of dietary fatty acid composition on muscle composition and hepatic fatty acid profile in juvenile Synechogobius hasta. J. Appl. Ichthyol. 2008, 24, 116–119. [Google Scholar] [CrossRef]
- Wen, Z.; Zhong, Z.; Darnell, J.E. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 1995, 82, 241–250. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Schindler, C. Regulation of Stat3 nuclear export. J. Clin. Investig. 2003, 111, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhang, T.; Novotny-Diermayr, V.; Tan, A.L.; Cao, X. A novel sequence in the coiled-coil domain of Stat3 essential for its nuclear translocation. J. Biol. Chem. 2003, 278, 29252–29260. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.S.; Ward, A.C. Conservation, duplication and divergence of the zebrafish STAT5 genes. Gene 2004, 338, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Schindler, C.; Darnell, J.E., Jr. Transcriptional responses to polypeptide ligands: The JAK-STAT pathway. Annu. Rev. Biochem. 1995, 64, 621–652. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; McBride, K.M.; Reich, N.C. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-α3. Proc. Natl. Acad. Sci. USA 2005, 102, 8150–8155. [Google Scholar] [CrossRef] [PubMed]
- Gunther, V.; Davis, A.M.; Georgiev, O.; Schaffner, W. A conserved cysteine cluster, essential for transcriptional activity, mediates homodimerization of human metal-responsive transcription factor-1 (MTF-1). Biochim. Biophys. Acta 2012, 1823, 476–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Gallego, M.I.; Smith, G.H.; Robinson, G.W.; Hennighausen, L. Functional rescue of Stat5a-null mammary tissue through the activation of compensating signals including STAT5B. Cell Growth Differ. 1998, 9, 795–804. [Google Scholar] [PubMed]
- Park, S.H.; Liu, X.; Hennighausen, L.; Davey, H.W.; Waxman, D.J. Distinctive roles of STAT5a and STAT5b in sexual dimorphism of hepatic P450 gene expression impact of Stat5a gene disruption. J. Biol. Chem. 1999, 274, 7421–7430. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.D.S.; Fernandes, M.N. Effect of copper on liver key enzymes of anaerobic glucose metabolism from freshwater tropical fish Prochilodus lineatus. Comp. Biochem. Physiol. A 2008, 151, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Kerner, J.; Hoppel, C. Fatty acid import into mitochondria. Biochim. Biophys. Acta 2000, 1486, 1–17. [Google Scholar] [CrossRef]
- Darnell, J.E. Reflections on STAT3, STAT5, and STAT6 as fat STATs. Proc. Natl. Acad. Sci. USA 1996, 93, 6221–6224. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.E.; Darnell, J.E. Stats: Transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002, 3, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Ghilardi, N.; Skoda, R.C. The leptin receptor activates Janus kinase 2 and signals for proliferation in a factor-dependent cell line. Mol. Endocrinol. 1997, 11, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Yeh, T.C.; Pellegrini, S. The Janus kinase family of protein tyrosine kinases and their role in signaling. Cell. Mol. Life Sci. 1999, 55, 1523–1534. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, L.A. The leptin receptor. J. Biol. Chem. 1997, 272, 6093–6096. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, K.; Saharinen, P.; Pesu, M.; Holt, V.E.; Silvennoinen, O.; O’Shea, J.J. The Janus kinases (Jaks). Genome Biol. 2004, 5, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Penn, O.; Privman, E.; Ashkenazy, H.; Landan, G.; Graur, D.; Pupko, T. GUIDANCE: A web server for assessing alignment confidence scores. Nucleic Acids Res. 2010, 38, W23–W28. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Liebel, S.; Oliveira Ribeiro, C.; Silva, R.; Ramsdorf, W.; Cestari, M.; Magalhaes, V.; Garcia, J.; Esquivel, B.; Filipak Neto, F. Cellular responses of Prochilodus lineatus hepatocytes after cylindrospermopsin exposure. Toxicol. Vitro 2011, 25, 1493–1500. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Zheng, J.L.; Luo, Z.; Chen, Q.L.; Zhu, Q.L. Carnitine palmitoyltransferase I gene in Synechogobius hasta: Cloning, mRNA expression and transcriptional regulation by insulin in vitro. Gene 2016, 576, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Hou, N.; Luo, M.S.; Liu, S.; Zhang, H.N.; Xiao, Q.; Sun, P.; Zhang, G.S.; Luo, J.D.; Chen, M.S. Leptin induceds hypertrophy through activating the peroxisome proliferator-activated receptor & pathway in cultured neonatal rat cardiomyocytes. Clin. Exp. Pharmacol. 2010, 37, 1087–1095. [Google Scholar]
- Zhuo, M.Q.; Luo, Z.; Wu, K.; Zhu, Q.L.; Zheng, J.L.; Chen, Q.L. Regulation of insulin on lipid metabolism in freshly isolated hepatocytes from yellow catfish (Pelteobagrus fulvidraco). Comp. Biochem. Physiol. B 2014, 177, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, K.; Leveille, G.A. Acetyl CoA carboxylase and fatty acid synthetase activities in liver and adipose tissue of meal-fed rats. Exp. Biol. Med. 1969, 131, 1051–1054. [Google Scholar] [CrossRef]
- Barroso, J.B.; Peragon, J.; Garcca-Salguero, L.; Dela-Higuera, M.; Lupianez, J.A. Variations in the kinetic behaviour of the NADPH-production systems in different tissues of the trout when fed on an amino-acid-based diet at different frequencies. Int. J. Biochem. Cell Biol. 1999, 31, 277–290. [Google Scholar] [CrossRef]
- Hisar, O.; Sonmez, A.Y.; Beydemir, S.; Hisar, S.A.; Yanik, T.; Cronin, T. Kinetic behavior of glucose 6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase in different tissues of rainbow trout (Oncorhynchus mykiss) exposed to non-lethal concentrations of cadmium. Acta Vet. Brno 2009, 78, 179–185. [Google Scholar] [CrossRef]
- Morash, A.J.; Kajimura, M.; McClelland, G.B. Intertissue regulation of carnitine palmitoyltransferase I (CPT I): Mitochondrial membrane properties and gene expression in rainbow trout (Oncorhynchus mykiss). Biochim. Biophys. Acta 2008, 1778, 1382–1389. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; Paepe, A.D.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]








| Gene | Accession No. | 5′ UTR (bp) | ORF (bp) | 3′ UTR (bp) | Full Length (bp) | Protein (aa) |
|---|---|---|---|---|---|---|
| STAT1-1 | KU532278 | 68 | 2292 | 915 | 3275 | 763 |
| STAT1-2 | KU532279 | 68 | 2253 | 915 | 3236 | 750 |
| STAT2 | KU532280 | 170 | 2484 | 1068 | 3722 | 827 |
| STAT3-1 | KU532281 | 224 | 2358 | 1090 | 3672 | 785 |
| STAT3-2 | KU532282 | 224 | 2298 | 1090 | 3612 | 765 |
| STAT4 | KU532283 | 146 | 2229 | 429 | 2804 | 742 |
| STAT5.1 | KU532284 | 180 | 2361 | 624 | 3165 | 786 |
| STAT5.2 | KU532285 | 125 | 2352 | 386 | 2863 | 783 |
| STAT6 | KU532286 | 462 | 2250 | 244 | 2956 | 749 |
| Species | STAT1 | STAT2 | STAT3 | STAT4 | STAT5.1 | STAT5.2 | STAT6 | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sh | Dr | Ga | Tr | Hs | Sh | Dr | Ga | Tr | Hs | Sh | Dr | Ga | Tr | Hs | Sh | Dr | Ga | Tr | Hs | Sh | Dr | Ga | Tr | Hs | Sh | Dr | Ga | Hs | Sh | Dr | Ga | Tr | Hs | |
| Sh S1 | *** | 61.5 | 80.4 | 80.7 | 67.7 | 42.2 | 43.3 | 44.1 | 44.7 | 39.4 | 50.4 | 49.4 | 50.3 | 51.0 | 50.5 | 48.3 | 51.3 | 49.7 | 48.9 | 49.2 | 27.4 | 27.3 | 27.8 | 27.7 | 27.1 | 26.7 | 27.3 | 26.4 | 27.0 | 24.2 | 25.1 | 24.5 | 23.7 | 23.0 |
| Dr S1 | – | *** | 63.3 | 62.5 | 61.9 | 42.7 | 42.5 | 42.4 | 42.7 | 37.6 | 47.3 | 47.4 | 47.8 | 47.7 | 47.9 | 46.7 | 49.8 | 48.7 | 47.7 | 48.7 | 28.0 | 28.2 | 27.9 | 27.7 | 27.6 | 27.4 | 27.0 | 26.7 | 26.7 | 26.0 | 27.0 | 25.2 | 25.0 | 25.2 |
| Ga S1 | *** | 83.8 | 67.6 | 41.3 | 42.5 | 42.9 | 44.4 | 39.7 | 50.3 | 50.0 | 50.5 | 50.9 | 51.1 | 47.4 | 50.1 | 48.6 | 48.8 | 48.7 | 26.7 | 26.6 | 26.9 | 26.8 | 26.7 | 26.1 | 26.2 | 25.8 | 26.4 | 24.0 | 25.4 | 24.4 | 23.9 | 23.1 | ||
| Tr S1 | *** | 67.7 | 40.9 | 41.5 | 42.4 | 43.6 | 39.3 | 49.7 | 48.8 | 49.3 | 49.9 | 50.0 | 47.9 | 51.2 | 49.0 | 48.8 | 49.5 | 26.3 | 26.5 | 26.7 | 26.7 | 26.6 | 25.5 | 26.2 | 25.6 | 26.0 | 24.8 | 25.2 | 24.0 | 23.3 | 22.0 | |||
| Hs S1 | *** | 44.2 | 46.4 | 45.3 | 46.1 | 42.3 | 51.4 | 51.2 | 51.6 | 52.2 | 51.8 | 53.1 | 53.6 | 53.3 | 52.3 | 53.3 | 28.0 | 28.0 | 27.9 | 27.8 | 28.6 | 27.4 | 28.3 | 27.0 | 28.4 | 26.8 | 26.7 | 26.2 | 25.3 | 23.7 | ||||
| Sh S2 | *** | 53.6 | 69.7 | 69.6 | 38.3 | 41.7 | 41.4 | 41.3 | 41.9 | 41.4 | 39.0 | 40.9 | 39.9 | 38.9 | 40.4 | 24.3 | 24.2 | 23.9 | 24.1 | 24.1 | 23.1 | 23.5 | 23.1 | 24.3 | 24.2 | 24.4 | 25.0 | 24.2 | 22.3 | |||||
| Dr S2 | *** | 56.9 | 58.5 | 36.3 | 42.7 | 41.7 | 42.1 | 43.0 | 42.9 | 39.8 | 40.9 | 39.7 | 39.2 | 41.5 | 27.1 | 27.0 | 27.3 | 27.4 | 26.2 | 27.2 | 26.7 | 27.2 | 26.4 | 25.0 | 25.3 | 26.1 | 24.3 | 22.6 | ||||||
| Ga S2 | *** | 74.5 | 43.1 | 43.2 | 43.0 | 43.2 | 43.5 | 43.2 | 39.5 | 41.1 | 40.1 | 39.1 | 41.1 | 25.3 | 25.4 | 26.0 | 25.9 | 26.5 | 24.6 | 25.4 | 24.9 | 26.3 | 24.9 | 25.4 | 25.8 | 24.6 | 23.2 | |||||||
| Tr S2 | *** | 41.9 | 44.3 | 44.6 | 44.3 | 44.6 | 44.1 | 40.9 | 41.7 | 40.7 | 40.9 | 41.8 | 26.8 | 26.6 | 26.8 | 26.5 | 27.0 | 26.4 | 27.0 | 26.6 | 27.0 | 25.7 | 26.2 | 26.4 | 26.3 | 24.8 | ||||||||
| Hs S2 | *** | 36.9 | 36.2 | 35.8 | 36.8 | 37.5 | 38.5 | 41.0 | 40.3 | 40.0 | 40.3 | 23.8 | 24.4 | 23.3 | 23.3 | 23.9 | 23.9 | 24.0 | 24.8 | 23.2 | 20.7 | 23.5 | 22.5 | 21.6 | 20.6 | |||||||||
| Sh S3 | *** | 95.7 | 97.1 | 96.3 | 88.7 | 47.5 | 48.6 | 47.9 | 47.1 | 48.5 | 29.1 | 29.5 | 29.5 | 29.3 | 29.7 | 29.3 | 29.2 | 29.5 | 30.0 | 22.7 | 25.9 | 24.9 | 23.8 | 22.0 | ||||||||||
| Dr S3 | *** | 92.3 | 94.5 | 88.5 | 47.4 | 48.9 | 48.1 | 47.5 | 48.7 | 28.5 | 29.2 | 29.1 | 28.8 | 28.8 | 28.9 | 28.7 | 29.1 | 28.9 | 22.5 | 25.0 | 24.9 | 23.5 | 21.2 | |||||||||||
| Ga S3 | *** | 95.3 | 87.9 | 47.9 | 49.0 | 48.0 | 47.3 | 49.2 | 28.4 | 29.1 | 29.0 | 28.7 | 29.2 | 28.5 | 28.4 | 28.9 | 29.3 | 22.7 | 25.2 | 25.1 | 23.2 | 21.6 | ||||||||||||
| Tr S3 | *** | 88.0 | 47.8 | 49.1 | 48.2 | 47.4 | 48.6 | 28.8 | 29.3 | 29.3 | 29.1 | 29.8 | 28.8 | 29.2 | 29.3 | 30.1 | 22.6 | 25.4 | 25.1 | 23.6 | 22.3 | |||||||||||||
| Hs S3 | *** | 47.0 | 48.8 | 47.9 | 46.8 | 48.2 | 28.2 | 28.6 | 28.7 | 28.6 | 28.6 | 28.3 | 28.9 | 28.7 | 28.8 | 22.4 | 25.2 | 25.0 | 24.0 | 22.6 | ||||||||||||||
| Sh S4 | *** | 73.2 | 86.0 | 81.7 | 59.1 | 27.0 | 28.0 | 27.7 | 27.5 | 28.4 | 27.7 | 27.9 | 27.9 | 28.5 | 25.8 | 25.7 | 25.4 | 25.9 | 25.8 | |||||||||||||||
| Dr S4 | *** | 74.8 | 72.4 | 61.0 | 28.6 | 28.9 | 28.8 | 28.6 | 28.9 | 27.8 | 27.9 | 27.9 | 28.6 | 25.1 | 26.6 | 25.4 | 26.4 | 26.4 | ||||||||||||||||
| Ga S4 | *** | 86.1 | 59.3 | 28.4 | 29.6 | 29.2 | 29.0 | 29.3 | 28.7 | 28.0 | 28.5 | 29.0 | 24.9 | 26.5 | 25.3 | 25.3 | 25.2 | |||||||||||||||||
| Tr S4 | *** | 58.3 | 27.7 | 28.3 | 28.2 | 27.9 | 28.4 | 28.3 | 27.9 | 28.0 | 28.3 | 25.0 | 26.4 | 25.4 | 25.5 | 25.3 | ||||||||||||||||||
| Hs S4 | *** | 29.4 | 29.8 | 29.5 | 29.2 | 29.8 | 28.8 | 28.9 | 28.7 | 29.5 | 24.8 | 25.6 | 25.3 | 25.9 | 24.1 | |||||||||||||||||||
| Sh S5.1 | *** | 92.6 | 94.3 | 94.9 | 79.1 | 81.0 | 82.7 | 81.5 | 78.0 | 34.9 | 34.6 | 39.0 | 34.9 | 36.7 | ||||||||||||||||||||
| Dr S5.1 | *** | 94.1 | 93.6 | 80.1 | 82.2 | 83.9 | 82.2 | 79.1 | 35.9 | 35.0 | 39.1 | 35.5 | 37.9 | |||||||||||||||||||||
| Ga S5.1 | *** | 96.3 | 79.6 | 82.3 | 82.8 | 82.0 | 78.2 | 35.3 | 34.4 | 39.4 | 35.2 | 36.9 | ||||||||||||||||||||||
| Tr S5.1 | *** | 79.9 | 82.7 | 83.2 | 82.5 | 78.7 | 35.5 | 34.8 | 39.5 | 35.5 | 37.7 | |||||||||||||||||||||||
| Hs S5B | *** | 74.2 | 74.2 | 74.8 | 93.7 | 34.6 | 33.9 | 37.9 | 35.4 | 37.8 | ||||||||||||||||||||||||
| Sh S5.2 | *** | 79.5 | 89.6 | 73.6 | 35.6 | 35.5 | 39.0 | 35.2 | 37.2 | |||||||||||||||||||||||||
| Dr S5.2 | *** | 78.6 | 74.5 | 35.5 | 35.6 | 38.6 | 35.1 | 37.0 | ||||||||||||||||||||||||||
| Ga S5.2 | *** | 74.0 | 34.2 | 34.0 | 38.0 | 35.5 | 37.0 | |||||||||||||||||||||||||||
| Hs S5A | *** | 34.6 | 34.1 | 38.2 | 35.2 | 37.7 | ||||||||||||||||||||||||||||
| Sh S6 | *** | 50.9 | 58.3 | 57.6 | 38.3 | |||||||||||||||||||||||||||||
| Dr S6 | *** | 58.7 | 53.7 | 37.3 | ||||||||||||||||||||||||||||||
| Ga S6 | *** | 74.4 | 41.4 | |||||||||||||||||||||||||||||||
| Tr S6 | *** | 37.4 | ||||||||||||||||||||||||||||||||
| Hs S6 | *** | |||||||||||||||||||||||||||||||||
| Values | 69.71 ± 2.72 | 54.24 ± 4.43 | 92.43 ± 1.20 | 71.19 ± 3.54 | 88.45 ± 2.41 | 78.30 ± 2.48 | 50.80 ± 3.85 | |||||||||||||||||||||||||||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, K.; Tan, X.-Y.; Wei, C.-C.; You, W.-J.; Zhuo, M.-Q.; Song, Y.-F. Isolation and Expression Analysis of STAT Members from Synechogobius hasta and Their Roles in Leptin Affecting Lipid Metabolism. Int. J. Mol. Sci. 2016, 17, 406. https://doi.org/10.3390/ijms17030406
Wu K, Tan X-Y, Wei C-C, You W-J, Zhuo M-Q, Song Y-F. Isolation and Expression Analysis of STAT Members from Synechogobius hasta and Their Roles in Leptin Affecting Lipid Metabolism. International Journal of Molecular Sciences. 2016; 17(3):406. https://doi.org/10.3390/ijms17030406
Chicago/Turabian StyleWu, Kun, Xiao-Ying Tan, Chuan-Chuan Wei, Wen-Jing You, Mei-Qin Zhuo, and Yu-Feng Song. 2016. "Isolation and Expression Analysis of STAT Members from Synechogobius hasta and Their Roles in Leptin Affecting Lipid Metabolism" International Journal of Molecular Sciences 17, no. 3: 406. https://doi.org/10.3390/ijms17030406
APA StyleWu, K., Tan, X. -Y., Wei, C. -C., You, W. -J., Zhuo, M. -Q., & Song, Y. -F. (2016). Isolation and Expression Analysis of STAT Members from Synechogobius hasta and Their Roles in Leptin Affecting Lipid Metabolism. International Journal of Molecular Sciences, 17(3), 406. https://doi.org/10.3390/ijms17030406