miRTargetLink—miRNAs, Genes and Interaction Networks
Abstract
:1. Introduction
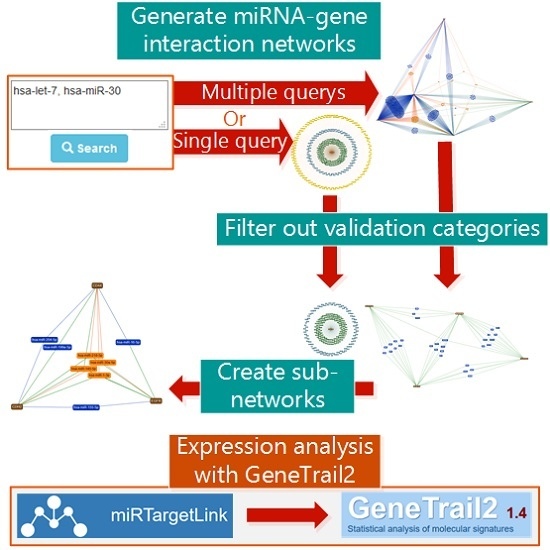
2. Results
3. Data Input
4. Constructing, Visualizing and Modifying Networks
5. Enrichment Analysis
6. Limitations and Future Research Directions
- (1)
- At this stage, the application is limited to Homo sapiens since most information is available and a considerable amount of biomedical research is carried out in humans. In the next release we will integrate other organisms, starting with Mus musculus and Rattus norvegicus. Along with more organisms, improved mapping functionality and support of other identifiers can facilitate the application of miRTargetLink.
- (2)
- Validated miRNA gene interactions are central for miRTargetLink. As a comprehensive resource for such interactions, we selected the miRTarBase. In addition to this database, many others are available (e.g., Tarbase or miRecords). To extend the tool beyond validated targets, predicted interactions are included. However, respective targets are known to depend on the prediction algorithm. Results can vary tremendously between different algorithms. In the present release, predicted interactions in miRTargetLink rely on miRanda. We have selected miRanda as a case of frequently applied prediction algorithms. Adding other prediction tools can potentially improve the results provided by miRTargetLink. Before adding those, the performance of the different methods, however, has to be evaluated critically to validate interactions.
- (3)
- The miRNA-gene predictions are known to be error-prone. This means that some of the miRNA-gene interactions that have been validated by life scientists may have been negative. The negative experiments are usually not reported. We are implementing a database for negative miRNA-gene interactions and will integrate the information in miRTargetLink so that potential users also get the information on false positive predictions.
7. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Snel, B.; Lehmann, G.; Bork, P.; Huynen, M.A. String: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000, 28, 3442–3444. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. String v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Laczny, C.; Leidinger, P.; Haas, J.; Ludwig, N.; Backes, C.; Gerasch, A.; Kaufmann, M.; Vogel, B.; Katus, H.A.; Meder, B.; et al. miRTrail—A comprehensive webserver for analyzing gene and miRNA patterns to enhance the understanding of regulatory mechanisms in diseases. BMC Bioinform. 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Hamed, M.; Spaniol, C.; Nazarieh, M.; Helms, V. TFmiR: A web server for constructing and analyzing disease-specific transcription factor and miRNA co-regulatory networks. Nucleic Acids Res. 2015, 43, W283–W288. [Google Scholar] [CrossRef] [PubMed]
- Moreira, F.C.; Dustan, B.; Hamoy, I.G.; Ribeiro-Dos-Santos, A.M.; Dos Santos, A.R. TargetCompare: A web interface to compare simultaneous miRNAs targets. Bioinformation 2014, 10, 602–605. [Google Scholar] [CrossRef] [PubMed]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.H.; Chang, N.W.; Shrestha, S.; Hsu, S.D.; Lin, Y.L.; Lee, W.H.; Yang, C.D.; Hong, H.C.; Wei, T.Y.; Tu, S.J.; et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016, 44, D239–D247. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5. [Google Scholar] [CrossRef] [PubMed]
- Backes, C.; Keller, A.; Kuentzer, J.; Kneissl, B.; Comtesse, N.; Elnakady, Y.A.; Muller, R.; Meese, E.; Lenhof, H.P. GeneTrail—Advanced gene set enrichment analysis. Nucleic Acids Res. 2007, 35, W186–W192. [Google Scholar] [CrossRef] [PubMed]
- miRTargetLink. Available online: http://ccb-web.cs.uni-saarland.de/mirtargetlink (accessed on 8 April 2016).
- VIS.js. Available online: http://www.visjs.org (accessed on 8 April 2016).
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- miEAA. Available online: www.ccb.uni-saarland.de/mieaa_tool/mirna_version_converter (accessed on 8 April 2016).
- Duan, L.; Xiong, X.; Liu, Y.; Wang, J. miRNA-1: Functional roles and dysregulation in heart disease. Mol. Biosyst. 2014, 10, 2775–2782. [Google Scholar] [CrossRef] [PubMed]
- Nabialek, E.; Wanha, W.; Kula, D.; Jadczyk, T.; Krajewska, M.; Kowalowka, A.; Dworowy, S.; Hrycek, E.; Wludarczyk, W.; Parma, Z.; et al. Circulating microRNAs (miR-423-5p, miR-208a and miR-1) in acute myocardial infarction and stable coronary heart disease. Minerva Cardioangiol. 2013, 61, 627–637. [Google Scholar] [PubMed]
- Higashi, K.; Yamada, Y.; Minatoguchi, S.; Baba, S.; Iwasa, M.; Kanamori, H.; Kawasaki, M.; Nishigaki, K.; Takemura, G.; Kumazaki, M.; et al. MicroRNA-145 repairs infarcted myocardium by accelerating cardiomyocyte autophagy. Am. J. Physiol. Heart Circ. Physiol. 2015, 309, H1813–H1826. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Du, G.Q.; Zhu, Z.T.; Zhang, C.; Sun, X.W.; Liu, J.J.; Li, X.; Wang, Y.S.; Du, W.J. Identification of apoptosis-related microRNAs and their target genes in myocardial infarction post-transplantation with skeletal myoblasts. J. Transl. Med. 2015, 13. [Google Scholar] [CrossRef] [PubMed]
- Stockel, D.; Kehl, T.; Trampert, P.; Schneider, L.; Backes, C.; Ludwig, N.; Gerasch, A.; Kaufmann, M.; Gessler, M.; Graf, N.; et al. Multi-omics Enrichment Analysis using the GeneTrail2 Web Service. Bioinformatics 2016, 32. [Google Scholar] [CrossRef] [PubMed]
- Backes, C.; Meese, E.; Lenhof, H.P.; Keller, A. A dictionary on microRNAs and their putative target pathways. Nucleic Acids Res. 2010, 38, 4476–4486. [Google Scholar] [CrossRef] [PubMed]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamberg, M.; Backes, C.; Fehlmann, T.; Hart, M.; Meder, B.; Meese, E.; Keller, A. miRTargetLink—miRNAs, Genes and Interaction Networks. Int. J. Mol. Sci. 2016, 17, 564. https://doi.org/10.3390/ijms17040564
Hamberg M, Backes C, Fehlmann T, Hart M, Meder B, Meese E, Keller A. miRTargetLink—miRNAs, Genes and Interaction Networks. International Journal of Molecular Sciences. 2016; 17(4):564. https://doi.org/10.3390/ijms17040564
Chicago/Turabian StyleHamberg, Maarten, Christina Backes, Tobias Fehlmann, Martin Hart, Benjamin Meder, Eckart Meese, and Andreas Keller. 2016. "miRTargetLink—miRNAs, Genes and Interaction Networks" International Journal of Molecular Sciences 17, no. 4: 564. https://doi.org/10.3390/ijms17040564
APA StyleHamberg, M., Backes, C., Fehlmann, T., Hart, M., Meder, B., Meese, E., & Keller, A. (2016). miRTargetLink—miRNAs, Genes and Interaction Networks. International Journal of Molecular Sciences, 17(4), 564. https://doi.org/10.3390/ijms17040564