Topoisomerase II Inhibitors Can Enhance Baculovirus-Mediated Gene Expression in Mammalian Cells through the DNA Damage Response
Abstract
:1. Introduction
2. Results
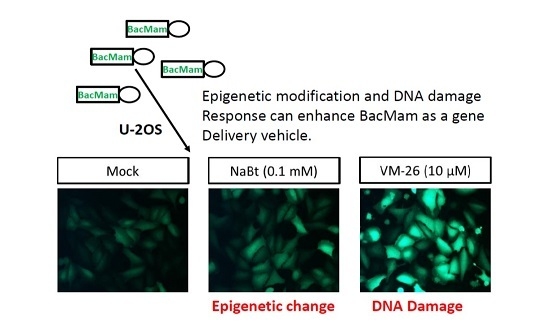
2.1. VM-26 Can Enhance Baculovirus-Transduced Gene Expression in U-2OS Cells
2.2. Top II but Not Top I Inhibitors Enhance Baculovirus-Delivered Gene Expression in U-2OS Cells
2.3. VM-26 Enhanced Baculovirus Gene Expression in U-2OS Cells through the DDR but Does Not Alter the Acetylation of Histone Proteins
2.4. Combined VM-26 with Baculovirus Mediated p53 Gene Therapy
3. Discussion
4. Materials and Methods
4.1. Cells
4.2. Construction of Plasmids
4.3. Recombinant Virus Production and Titer Determination
4.4. Western Blot Analysis
4.5. Transduction of Mammalian Cells
4.6. EGFP Expression Analysis
4.7. Cell Viability Assay
4.8. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Herniou, E.A.; Jehle, J.A. Baculovirus phylogeny and evolution. Curr. Drug Targets 2007, 8, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Van Oers, M.M.; Pijlman, G.P.; Vlak, J.M. Thirty years of baculovirus-insect cell protein expression: From dark horse to mainstream technology. J. Gen. Virol. 2015, 96, 6–23. [Google Scholar] [CrossRef] [PubMed]
- Ayres, M.D.; Howard, S.C.; Kuzio, J.; Lopez-Ferber, M.; Possee, R.D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 1994, 202, 586–605. [Google Scholar] [CrossRef] [PubMed]
- Herniou, E.A.; Olszewski, J.A.; Cory, J.S.; O’Reilly, D.R. The genome sequence and evolution of baculoviruses. Annu. Rev. Entomol. 2003, 48, 211–234. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.E.; Summers, M.D.; Fraser, M.J. Production of human β interferon in insect cells infected with a baculovirus expression vector. Mol. Cell. Biol. 1983, 3, 2156. [Google Scholar] [CrossRef] [PubMed]
- Kost, T.A.; Condreay, J.P.; Jarvis, D.L. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat. Biotechnol. 2005, 23, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Deschuyteneer, M.; Elouahabi, A.; Plainchamp, D.; Plisnier, M.; Soete, D.; Corazza, Y.; Lockman, L.; Giannini, S.; Deschamps, M. Molecular and structural characterization of the L1 virus-like particles that are used as vaccine antigens in Cervarix™, the AS04-adjuvanted HPV-16 and -18 cervical cancer vaccine. Hum. Vaccines 2010, 6, 407–419. [Google Scholar] [CrossRef]
- Huber, V.C.; McCullers, J.A. FluBlok, a recombinant influenza vaccine. Curr. Opin. Mol. Ther. 2008, 10, 75–85. [Google Scholar] [PubMed]
- Kost, T.A.; Condreay, J.P.; Ames, R.S. Baculovirus gene delivery: A flexible assay development tool. Curr. Gene Ther. 2010, 10, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, C.; Sandig, V.; Jennings, G.; Rudolph, M.; Schlag, P.; Strauss, M. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc. Natl. Acad. Sci. USA 1995, 92, 10099–10103. [Google Scholar] [CrossRef] [PubMed]
- Boyce, F.M.; Bucher, N.L. Baculovirus-mediated gene transfer into mammalian cells. Proc. Natl. Acad. Sci. USA 1996, 93, 2348–2352. [Google Scholar] [CrossRef] [PubMed]
- Shoji, I.; Aizaki, H.; Tani, H.; Ishii, K.; Chiba, T.; Saito, I.; Miyamura, T.; Matsuura, Y. Efficient gene transfer into various mammalian cells, including non-hepatic cells, by baculovirus vectors. J. Gen. Virol. 1997, 78, 2657–2664. [Google Scholar] [CrossRef] [PubMed]
- Wagle, M.; Jesuthasan, S. Baculovirus-mediated gene expression in zebrafish. Mar. Biotechnol. (NY) 2003, 5, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Leisy, D.J.; Lewis, T.D.; Leong, J.-A.C.; Rohrmann, G.F. Transduction of cultured fish cells with recombinant baculoviruses. J. Gen. Virol. 2003, 84, 1173–1178. [Google Scholar] [CrossRef] [PubMed]
- Ames, R.S.; Kost, T.A.; Condreay, J.P. BacMam technology and its application to drug discovery. Expert Opin. Drug Discov. 2007, 2, 1669–1681. [Google Scholar] [CrossRef] [PubMed]
- Airenne, K.J.; Hu, Y.-C.; Kost, T.A.; Smith, R.H.; Kotin, R.M.; Ono, C.; Matsuura, Y.; Wang, S.; Ylä-Herttuala, S. Baculovirus: An insect-derived vector for diverse gene transfer applications. Mol. Ther. 2013, 21, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Ames, R.; Fornwald, J.; Nuthulaganti, P.; Trill, J.; Foley, J.; Buckley, P.; Kost, T.; Wu, Z.; Romanos, M. BacMam recombinant baculoviruses in G protein-coupled receptor drug discovery. Recept. Channels 2004, 10, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Choudhury, Y.; Yang, J.; Chen, C.; Tay, F.C.; Lim, T.M.; Wang, S. Antiglioma effects of combined use of a baculovirual vector expressing wild-type p53 and sodium butyrate. J. Gene Med. 2011, 13, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-L.; Luo, W.-Y.; Lo, W.-H.; Lin, K.-J.; Sung, L.-Y.; Shih, Y.-S.; Chang, Y.-H.; Hu, Y.-C. Development of hybrid baculovirus vectors for artificial microRNA delivery and prolonged gene suppression. Biotechnol. Bioeng. 2011, 108, 2958–2967. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-L.; Wu, J.-C.; Chen, G.-Y.; Yuan, P.-H.; Tseng, Y.-W.; Li, K.-C.; Hwang, S.-M.; Hu, Y.-C. Baculovirus-mediated miRNA regulation to suppress hepatocellular carcinoma tumorigenicity and metastasis. Mol. Ther. 2015, 23, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Lau, C.-H.; Goh, S.-L.; Liang, Q.; Chen, C.; Du, S.; Phang, R.-Z.; Tay, F.C.; Tan, W.-K.; Li, Z.; et al. Baculoviral transduction facilitates TALEN-mediated targeted transgene integration and Cre/LoxP cassette exchange in human-induced pluripotent stem cells. Nucleic Acids Res. 2013, 41. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Z.; Wu, C.P.; Chao, Y.-C.; Liu, C.Y.-Y. Membrane penetrating peptides greatly enhance baculovirus transduction efficiency into mammalian cells. Biochem. Biophys. Res. Commun. 2011, 405, 297–302. [Google Scholar] [CrossRef]
- Laakkonen, J.P.; Mäkelä, A.R.; Kakkonen, E.; Turkki, P.; Kukkonen, S.; Peränen, J.; Ylä-Herttuala, S.; Airenne, K.J.; Oker-Blom, C.; Vihinen-Ranta, M.; et al. Clathrin-independent entry of baculovirus triggers uptake of E. coli in non-phagocytic human cells. PLoS ONE 2009, 4, e5093. [Google Scholar] [CrossRef]
- Long, G.; Pan, X.; Kormelink, R.; Vlak, J.M. Functional entry of baculovirus into insect and mammalian cells is dependent on clathrin-mediated endocytosis. J. Virol. 2006, 80, 8830–8833. [Google Scholar] [CrossRef] [PubMed]
- Condreay, J.P.; Witherspoon, S.M.; Clay, W.C.; Kost, T.A. Transient and stable gene expression in mammalian cells transduced with a recombinant baculovirus vector. Proc. Natl. Acad. Sci. USA 1999, 96, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Halazonetis, T.D.; Gorgoulis, V.G.; Bartek, J. An oncogene-induced DNA damage model for cancer development. Science 2008, 319, 1352–1355. [Google Scholar] [CrossRef] [PubMed]
- Xiaofei, E.; Kowalik, T.F. The DNA damage response induced by infection with human cytomegalovirus and other viruses. Viruses 2014, 6, 2155–2185. [Google Scholar] [PubMed]
- Smith, S.; Weller, S.K. HSV-I and the cellular DNA damage response. Future Virol. 2015, 10, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.K.; Byers, N.M.; Friesen, P.D. Baculovirus F-box protein LEF-7 modifies the host DNA damage response to enhance virus multiplication. J. Virol. 2013, 87, 12592–12599. [Google Scholar] [CrossRef] [PubMed]
- Hsiang, Y.H.; Hertzberg, R.; Hecht, S.; Liu, L.F. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 1985, 260, 14873–14878. [Google Scholar] [PubMed]
- Liu, L.F. DNA topoisomerase poisons as antitumor drugs. Annu. Rev. Biochem. 1989, 58, 351–375. [Google Scholar] [CrossRef] [PubMed]
- Spenger, A.; Ernst, W.; Condreay, J.P.; Kost, T.A.; Grabherr, R. Influence of promoter choice and trichostatin A treatment on expression of baculovirus delivered genes in mammalian cells. Protein Expr. Purif. 2004, 38, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Mao, Y.; Desai, S.D.; Zhou, N.; Ting, C.-Y.; Hwang, J.; Liu, L.F. The topoisomerase IIβ circular clamp arrests transcription and signals a 26S proteasome pathway. Proc. Natl. Acad. Sci. USA. 2003, 100, 3239–3244. [Google Scholar] [CrossRef] [PubMed]
- Song, S.U.; Boyce, F.M. Combination treatment for osteosarcoma with baculoviral vector mediated gene therapy (p53) and chemotherapy (Adriamycin). Exp. Mol. Med. 2001, 33, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.-K.; Lin, J.-Z.; Jinn, T.-R.; Chan, H.-L.; Wu, T.-Y. Identification of Rhopalosiphum padi virus 5′ untranslated region sequences required for cryptic promoter activity and internal ribosome entry. Int. J. Mol. Sci. 2015, 16, 16053–16066. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.-Y.; Liu, M.-K. Chimeral Internal Ribosomal Entry Site Sequence and Uses Thereof, 2013. Available online: http://www.google.com/patents/US20120231536 (accessed on 5 February 2013).
- Rivera-Gonzalez, G.C.; Swift, S.L.; Dussupt, V.; Georgopoulos, L.J.; Maitland, N.J. Baculoviruses as gene therapy vectors for human prostate cancer. J. Invertebr. Pathol. 2011, 107 (Suppl.), S59–S70. [Google Scholar] [CrossRef] [PubMed]
- Swift, S.L.; Rivera, G.C.; Dussupt, V.; Leadley, R.M.; Hudson, L.C.; Ma de Ridder, C.; Kraaij, R.; Burns, J.E.; Maitland, N.J.; Georgopoulos, L.J. Evaluating baculovirus as a vector for human prostate cancer gene therapy. PLoS ONE 2013, 8, e65557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandig, V.; Hofmann, C.; Steinert, S.; Jennings, G.; Schlag, P.; Strauss, M. Gene transfer into hepatocytes and human liver tissue by baculovirus vectors. Hum. Gene Ther. 1996, 7, 1937–1945. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-L.; Tseng, Y.-W.; Wu, J.-C.; Chen, G.-Y.; Lin, K.-C.; Hwang, S.-M.; Hu, Y.-C. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and microRNA regulation. Biomaterials 2015, 44, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Sung, L.-Y.; Chen, C.-L.; Lin, S.-Y.; Li, K.-C.; Yeh, C.-L.; Chen, G.-Y.; Lin, C.-Y.; Hu, Y.-C. Efficient gene delivery into cell lines and stem cells using baculovirus. Nat. Protoc. 2014, 9, 1882–1899. [Google Scholar] [CrossRef] [PubMed]
- Nevels, M.; Nitzsche, A.; Paulus, C. How to control an infectious bead string: Nucleosome-based regulation and targeting of herpesvirus chromatin. Rev. Med. Virol. 2011, 21, 154–180. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Reuven, N.; Mohni, K.N.; Schumacher, A.J.; Weller, S.K. Structure of the herpes simplex virus 1 genome: Manipulation of nicks and gaps can abrogate infectivity and alter the cellular DNA damage response. J. Virol. 2014, 88, 10146–10156. [Google Scholar] [CrossRef] [PubMed]
- Volcy, K.; Fraser, N.W. DNA damage promotes herpes simplex virus-1 protein expression in a neuroblastoma cell line. J. Neurovirol. 2013, 19, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Stanulla, M.; Wang, J.; Chervinsky, D.S.; Aplan, P.D. Topoisomerase II inhibitors induce DNA double-strand breaks at a specific site within the AML1 locus. Leukemia 1997, 11, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-J.; Chen, W.-S.; Wu, T.-Y. Development of a bi-cistronic baculovirus expression vector by the Rhopalosiphum padi virus 5′ internal ribosome entry site. Biochem. Biophys. Res. Commun. 2005, 335, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Keil, G.M.; Klopfleisch, C.; Giesow, K.; Blohm, U. Novel vectors for simultaneous high-level dual protein expression in vertebrate and insect cells by recombinant baculoviruses. J. Virol. Methods 2009, 160, 132–137. [Google Scholar] [CrossRef] [PubMed]








© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.-K.; Lin, J.-J.; Chen, C.-Y.; Kuo, S.-C.; Wang, Y.-M.; Chan, H.-L.; Wu, T.Y. Topoisomerase II Inhibitors Can Enhance Baculovirus-Mediated Gene Expression in Mammalian Cells through the DNA Damage Response. Int. J. Mol. Sci. 2016, 17, 931. https://doi.org/10.3390/ijms17060931
Liu M-K, Lin J-J, Chen C-Y, Kuo S-C, Wang Y-M, Chan H-L, Wu TY. Topoisomerase II Inhibitors Can Enhance Baculovirus-Mediated Gene Expression in Mammalian Cells through the DNA Damage Response. International Journal of Molecular Sciences. 2016; 17(6):931. https://doi.org/10.3390/ijms17060931
Chicago/Turabian StyleLiu, Ming-Kun, Jhe-Jhih Lin, Chung-Yung Chen, Szu-Cheng Kuo, Yu-Ming Wang, Hong-Lin Chan, and Tzong Yuan Wu. 2016. "Topoisomerase II Inhibitors Can Enhance Baculovirus-Mediated Gene Expression in Mammalian Cells through the DNA Damage Response" International Journal of Molecular Sciences 17, no. 6: 931. https://doi.org/10.3390/ijms17060931
APA StyleLiu, M.-K., Lin, J.-J., Chen, C.-Y., Kuo, S.-C., Wang, Y.-M., Chan, H.-L., & Wu, T. Y. (2016). Topoisomerase II Inhibitors Can Enhance Baculovirus-Mediated Gene Expression in Mammalian Cells through the DNA Damage Response. International Journal of Molecular Sciences, 17(6), 931. https://doi.org/10.3390/ijms17060931