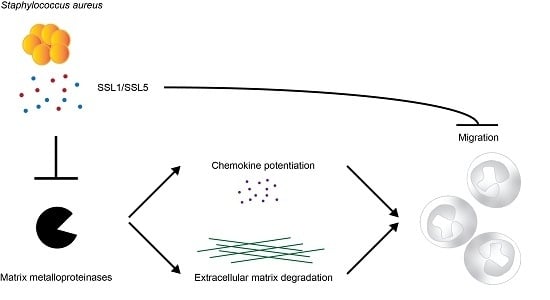
Staphylococcal Superantigen-Like Protein 1 and 5 (SSL1 & SSL5) Limit Neutrophil Chemotaxis and Migration through MMP-Inhibition
Abstract
:1. Introduction
2. Results
2.1. Identification of Two Staphylococcal Inhibitors of Neutrophil Matrix Metalloproteinases (MMPs)
2.2. Staphylococcal Superantigen-Like Protein 1 and 5 (SSL1 and SSL5) are Broad-Range MMP Inhibitors
2.3. The Potentiation of Neutrophil-Attracting Chemokines Is Inhibited by SSL1 and SSL5
2.4. SSL1 and SSL5 Inhibit Neutrophil Migration through Collagen
3. Discussion
4. Materials and Methods
4.1. Reagents and Chemicals
4.2. Cloning, Expression and Purification of Recombinant Staphylococcal Proteins
4.3. Cells
4.4. Trypsin Activation of the MMPs
4.5. Fluorogenic Peptide MMP Activity Assay
4.6. Far Western Blot to Detect MMP-SSL Binding
4.7. MMP-SSL Binding Enzyme-Linked Immunosorbent Assay (ELISA)
4.8. Western Blot to Visualize IL-8 Cleavage
4.9. Calcium Mobilization Assay
4.10. Visualization of MMP-Mediated Collagen Degradation
4.11. Neutrophil Migration Assays
4.12. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Nagase, H.; Visse, R.; Murphy, G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res. 2006, 69, 562–573. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, D.; Morrison, C.J.; Overall, C.M. Matrix metalloproteinases: What do they not do? New substrates and biological roles identified by murine models and proteomics. Biochim. Biophys. Acta 2010, 1803, 39–54. [Google Scholar] [CrossRef] [PubMed]
- Parks, W.C.; Wilson, C.L.; López-Boado, Y.S. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat. Rev. Immunol. 2004, 4, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Van Lint, P.; Libert, C. Chemokine and cytokine processing by matrix metalloproteinases and its effect on leukocyte migration and inflammation. J. Leukoc. Biol. 2007, 82, 1375–1381. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.; Albrecht, S.; Märki, C. Proteolytic processing of chemokines: Implications in physiological and pathological conditions. Int. J. Biochem. Cell Biol. 2008, 40, 1185–1198. [Google Scholar] [CrossRef] [PubMed]
- Koymans, K.J.; Vrieling, M.; Gorham, R.D.; van Strijp, J.A.G. Staphylococcal immune evasion proteins: Structure, function, and host adaptation. Curr. Top. Microbiol. Immunol. Available online: http://link.springer.com/chapter/10.1007/82_2015_5017 (accessed on 7 January 2016).
- Van Kessel, K.P.M.; Bestebroer, J.; van Strijp, J.A.G. Neutrophil-mediated phagocytosis of Staphylococcus aureus. Front. Immunol. 2014, 5, 467. [Google Scholar] [CrossRef] [PubMed]
- Postma, B.; Poppelier, M.J.; van Galen, J.C.; Prossnitz, E.R.; van Strijp, J.A.G.; de Haas, C.J.C.; van Kessel, K.P.M. Chemotaxis inhibitory protein of Staphylococcus aureus binds specifically to the C5a and formylated peptide receptor. J. Immunol. 2004, 172, 6994–7001. [Google Scholar] [CrossRef] [PubMed]
- Bestebroer, J.; Poppelier, M.J.J.G.; Ulfman, L.H.; Lenting, P.J.; Denis, C.V.; van Kessel, K.P.M.; van Strijp, J.A.G.; de Haas, C.J.C. Staphylococcal superantigen-like 5 binds PSGL-1 and inhibits P-selectin-mediated neutrophil rolling. Blood 2007, 109, 2936–2943. [Google Scholar] [CrossRef] [PubMed]
- Itoh, S.; Hamada, E.; Kamoshida, G.; Takeshita, K.; Oku, T.; Tsuji, T. Staphylococcal superantigen-like protein 5 inhibits matrix metalloproteinase 9 from human neutrophils. Infect. Immun. 2010, 78, 3298–3305. [Google Scholar] [CrossRef] [PubMed]
- Tester, A.M.; Cox, J.H.; Connor, A.R.; Starr, A.E.; Dean, R.A.; Puente, X.S.; López-Otín, C.; Overall, C.M. LPS responsiveness and neutrophil chemotaxis in vivo require PMN MMP-8 activity. PLoS ONE 2007, 2, e312. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Jackson, P.; Tester, A.M.; Diaconu, E.; Overall, C.M.; Blalock, J.E.; Pearlman, E. Matrix metalloproteinase-8 facilitates neutrophil migration through the corneal stromal matrix by collagen degradation and production of the chemotactic peptide Pro–Gly–Pro. Am. J. Pathol. 2008, 173, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Van den Steen, P.E.; Proost, P.; Wuyts, A.; van Damme, J.; Opdenakker, G. Neutrophil gelatinase B potentiates interleukin-8 tenfold by aminoterminal processing, whereas it degrades CTAP-III, PF-4, and GRO-α and leaves RANTES and MCP-2 intact. Blood 2000, 96, 2673–2681. [Google Scholar] [PubMed]
- Van Den Steen, P.E.; Wuyts, A.; Husson, S.J.; Proost, P.; van Damme, J.; Opdenakker, G. Gelatinase B/MMP-9 and neutrophil collagenase/MMP-8 process the chemokines human GCP-2/CXCL6, ENA-78/CXCL5 and mouse GCP-2/LIX and modulate their physiological activities. Eur. J. Biochem. 2003, 270, 3739–3749. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wu, C.; Zhang, X.; Sorokin, L.M. In vivo processing of CXCL5 (LIX) by matrix metalloproteinase (MMP)-2 and MMP-9 promotes early neutrophil recruitment in IL-1β-induced peritonitis. J. Immunol. 2013, 190, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.S.; Guo, Y.; Ramos, R.I.; Hebroni, F.; Plaisier, S.B.; Xuan, C.; Granick, J.L.; Matsushima, H.; Takashima, A.; Iwakura, Y.; et al. Neutrophil-derived IL-1β is sufficient for abscess formation in immunity against Staphylococcus aureus in mice. PLoS Pathog. 2012, 8, e1003047. [Google Scholar] [CrossRef] [PubMed]
- Schönbeck, U.; Mach, F.; Libby, P. Generation of biologically active IL-1 beta by matrix metalloproteinases: A novel caspase-1-independent pathway of IL-1 beta processing. J. Immunol. 1998, 161, 3340–3346. [Google Scholar] [PubMed]
- Overbeek, S.A.; Henricks, P.A.J.; Srienc, A.I.; Koelink, P.J.; de Kruijf, P.; Lim, H.D.; Smit, M.J.; Zaman, G.J.R.; Garssen, J.; Nijkamp, F.P.; et al. N-Acetylated Proline–Glycine–Proline induced G-protein dependent chemotaxis of neutrophils is independent of CXCL8 release. Eur. J. Pharmacol. 2011, 668, 428–434. [Google Scholar] [CrossRef] [PubMed]
- De Ceuninck, F.; Allain, F.; Caliez, A.; Spik, G.; Vanhoutte, P.M. High binding capacity of cyclophilin B to chondrocyte heparan sulfate proteoglycans and its release from the cell surface by matrix metalloproteinases: Possible role as a proinflammatory mediator in arthritis. Arthritis Rheum. 2003, 48, 2197–2206. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.L.; Ouellette, A.J.; Satchell, D.P.; Ayabe, T.; López-Boado, Y.S.; Stratman, J.L.; Hultgren, S.J.; Matrisian, L.M.; Parks, W.C. Regulation of intestinal α-defensin activation by the metalloproteinase matrilysin in innate host defense. Science 1999, 286, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Hartzell, W.; Shapiro, S.D. Macrophage elastase prevents Gemella morbillorum infection and improves outcome following murine bone marrow transplantation. Chest 1999, 116, 31S–32S. [Google Scholar] [CrossRef] [PubMed]
- Bestebroer, J.; van Kessel, K.P.M.; Azouagh, H.; Walenkamp, A.M.; Boer, I.G.J.; Romijn, R.A.; van Strijp, J.A.G.; de Haas, C.J.C. Staphylococcal SSL5 inhibits leukocyte activation by chemokines and anaphylatoxins. Blood 2009, 113, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Calander, A.-M.; Starckx, S.; Opdenakker, G.; Bergin, P.; Quiding-Järbrink, M.; Tarkowski, A. Matrix metalloproteinase-9 (gelatinase B) deficiency leads to increased severity of Staphylococcus aureus-triggered septic arthritis. Microbes Infect. 2006, 8, 1434–1439. [Google Scholar] [CrossRef] [PubMed]
- Kanangat, S.; Postlethwaite, A.; Hasty, K.; Kang, A.; Smeltzer, M.; Appling, W.; Schaberg, D. Induction of multiple matrix metalloproteinases in human dermal and synovial fibroblasts by Staphylococcus aureus: Implications in the pathogenesis of septic arthritis and other soft tissue infections. Arthritis Res. Ther. 2006, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.E.; Pettersen, S.; Stuestol, J.F.; Wang, Y.Y.; Foster, S.J.; Thiemermann, C.; Aasen, A.O.; Bjørnland, K. Peptidoglycan of S. aureus causes increased levels of matrix metalloproteinases in the rat. Shock 2004, 22, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Myhre, A.E.; Pettersen, S.J.; Dahle, M.K.; Foster, S.J.; Thiemermann, C.; Bjørnland, K.; Aasen, A.O.; Wang, J.E. Peptidoglycan of Staphylococcus aureus induces enhanced levels of matrix metalloproteinase-9 in human blood originating from neutrophils. Shock 2005, 24, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, H.-L.; Lin, C.-C.; Shih, R.-H.; Hsiao, L.-D.; Yang, C.-M. NADPH oxidase-mediated redox signal contributes to lipoteichoic acid-induced MMP-9 upregulation in brain astrocytes. J. Neuroinflamm. 2012, 9, 110. [Google Scholar] [CrossRef] [PubMed]
- Santala, A.; Saarinen, J.; Kovanen, P.; Kuusela, P. Activation of interstitial collagenase, MMP-1, by Staphylococcus aureus cells having surface-bound plasmin: A novel role of plasminogen receptors of bacteria. FEBS Lett. 1999, 461, 153–156. [Google Scholar] [CrossRef]
- Starr, A.E.; Dufour, A.; Maier, J.; Overall, C.M. Biochemical analysis of matrix metalloproteinase activation of chemokines CCL15 and CCL23 and increased glycosaminoglycan binding of CCL16. J. Biol. Chem. 2012, 287, 5848–5860. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.M.; Gaggar, A.; Blalock, J.E. MMP generated matrikines. Matrix Biol. 2015, 44, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Jackson, P.L.; Tanner, S.; Hardison, M.T.; Abdul Roda, M.; Blalock, J.E.; Gaggar, A. A self-propagating matrix metalloprotease-9 (MMP-9) dependent cycle of chronic neutrophilic inflammation. PLoS ONE 2011, 6, e15781. [Google Scholar] [CrossRef] [PubMed]
- Brew, K.; Nagase, H. The tissue inhibitors of metalloproteinases (TIMPs): An ancient family with structural and functional diversity. Biochim. Biophys. Acta 2010, 1803, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Bardoel, B.; Vos, R.; Bouman, T. Evasion of Toll-like receptor 2 activation by staphylococcal superantigen-like protein 3. J. Mol. Med. 2012, 90, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.C.; Wines, B.D.; Baker, H.; Langley, R.J.; Baker, E.N.; Fraser, J.D. The crystal structure of staphylococcal superantigen-like protein 11 in complex with sialyl Lewis X reveals the mechanism for cell binding and immune inhibition. Mol. Microbiol. 2007, 66, 1342–1355. [Google Scholar] [CrossRef] [PubMed]
- Baker, H.M.; Basu, I.; Chung, M.C.; Caradoc-Davies, T.; Fraser, J.D.; Baker, E.N. Crystal structures of the staphylococcal toxin SSL5 in complex with sialyl Lewis X reveal a conserved binding site that shares common features with viral and bacterial sialic acid binding proteins. J. Mol. Biol. 2007, 374, 1298–1308. [Google Scholar] [CrossRef] [PubMed]
- Koymans, K.J.; Feitsma, L.J.; Brondijk, T.H.C.; Aerts, P.C.; Lukkien, E.; Lössl, P.; van Kessel, K.P.M.; de Haas, C.J.C.; van Strijp, J.A.G.; Huizinga, E.G. Structural basis for inhibition of TLR2 by staphylococcal superantigen-like protein 3 (SSL3). Proc. Natl. Acad. Sci. USA 2015, 112, 11018–11023. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, R.; Itoh, S.; Kamoshida, G.; Takii, T.; Fujii, S.; Tsuji, T.; Onozaki, K. Staphylococcal superantigen-like protein 3 binds to the toll-like receptor 2 extracellular domain and inhibits cytokine production induced by Staphylococcus aureus, cell wall component, or lipopeptides in murine macrophages. Infect. Immun. 2012, 80, 2816–2825. [Google Scholar] [CrossRef] [PubMed]
- Laursen, N.S.; Gordon, N.; Hermans, S.; Lorenz, N.; Jackson, N.; Wines, B.; Spillner, E.; Christensen, J.B.; Jensen, M.; Fredslund, F.; et al. Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2010, 107, 3681–3686. [Google Scholar] [CrossRef] [PubMed]
- Gjertsson, I.; Innocenti, M.; Matrisian, L.M.; Tarkowski, A. Metalloproteinase-7 contributes to joint destruction in Staphylococcus aureus induced arthritis. Microb. Pathog. 2005, 38, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Elkington, P.T.G.; O’Kane, C.M.; Friedland, J.S. The paradox of matrix metalloproteinases in infectious disease. Clin. Exp. Immunol. 2005, 142, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Van de Weijer, M.L.; Bassik, M.C.; Luteijn, R.D.; Voorburg, C.M.; Lohuis, M.A.M.; Kremmer, E.; Hoeben, R.C.; LeProust, E.M.; Chen, S.; Hoelen, H.; et al. A high-coverage shRNA screen identifies TMEM129 as an E3 ligase involved in ER-associated protein degradation. Nat. Commun. 2014, 5, 3832. [Google Scholar] [CrossRef] [PubMed]





© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koymans, K.J.; Bisschop, A.; Vughs, M.M.; Van Kessel, K.P.M.; De Haas, C.J.C.; Van Strijp, J.A.G. Staphylococcal Superantigen-Like Protein 1 and 5 (SSL1 & SSL5) Limit Neutrophil Chemotaxis and Migration through MMP-Inhibition. Int. J. Mol. Sci. 2016, 17, 1072. https://doi.org/10.3390/ijms17071072
Koymans KJ, Bisschop A, Vughs MM, Van Kessel KPM, De Haas CJC, Van Strijp JAG. Staphylococcal Superantigen-Like Protein 1 and 5 (SSL1 & SSL5) Limit Neutrophil Chemotaxis and Migration through MMP-Inhibition. International Journal of Molecular Sciences. 2016; 17(7):1072. https://doi.org/10.3390/ijms17071072
Chicago/Turabian StyleKoymans, Kirsten J., Adinda Bisschop, Mignon M. Vughs, Kok P. M. Van Kessel, Carla J. C. De Haas, and Jos A. G. Van Strijp. 2016. "Staphylococcal Superantigen-Like Protein 1 and 5 (SSL1 & SSL5) Limit Neutrophil Chemotaxis and Migration through MMP-Inhibition" International Journal of Molecular Sciences 17, no. 7: 1072. https://doi.org/10.3390/ijms17071072
APA StyleKoymans, K. J., Bisschop, A., Vughs, M. M., Van Kessel, K. P. M., De Haas, C. J. C., & Van Strijp, J. A. G. (2016). Staphylococcal Superantigen-Like Protein 1 and 5 (SSL1 & SSL5) Limit Neutrophil Chemotaxis and Migration through MMP-Inhibition. International Journal of Molecular Sciences, 17(7), 1072. https://doi.org/10.3390/ijms17071072