The Profiling and Identification of the Absorbed Constituents and Metabolites of Guizhi Decoction in Rat Plasma and Urine by Rapid Resolution Liquid Chromatography Combined with Quadrupole-Time-of-Flight Mass Spectrometry
Abstract
:1. Introduction
2. Results
2.1. Analysis the Constituents of GZD
2.2. Analysis of Prototype Compounds in Plasma Sample
2.3. Analysis of Metabolites in Blood and Urine Samples
2.3.1. Characterization of (epi) Catechin-Related Metabolites
2.3.2. Characterization of Gallic Acid-Related Metabolites
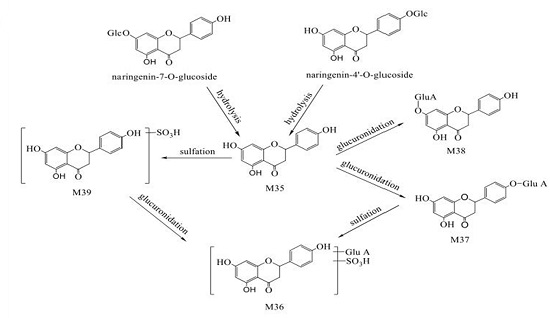
2.3.3. Characterization of Flavone-Related Metabolites
2.3.4. Characterization of Other Metabolites
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Rapid Resolution Liquid Chromatography with Quadrupole-Time-of-Flight Mass Spectrometry (RRLC-Q-TOF-MS) Analysis
4.3. Preparation of Guizhi Decoction (GZD)
4.4. Animal Housing Environment and Experiments
4.5. Sample Preparation
4.5.1. Preparation of GZD
4.5.2. Preparation of Plasma Samples
4.5.3. Preparation of Urine Samples
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Liu, S.H.; Chuang, W.C.; Lam, W. Safety surveillance of traditional Chinese medicine: Current and future. Drug Saf. 2015, 38, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.Y.; Chang, H.H.; Wu, M.Y.; Lin, H.C. Yin-Chen-Hao-Tang ameliorates obstruction-induced hepatic apoptosis in rats. J. Pharm. Pharmacol. 2007, 59, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Feng, F. Identification of components in Zhi-Zi-Da-Huang decoction by HPLC coupled with electrospray ionization tandem mass spectrometry, photodiode array and fluorescence detectors. J. Pharm. Biomed. Anal. 2009, 49, 1157–1165. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.Y. Therapeutic wisdom in traditional Chinese medicine: A perspective from modern science. Trends Pharmacol. Sci. 2005, 26, 558–563. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhou, T.Y.; Chen, M.J.; Jiang, M.; Wang, X.X.; Liu, Q.; Zhan, Z.; Zhang, X. Research progress on synergistic anti-tumor mechanisms of compounds in traditional Chinese medicine. J. Tradit. Chin. Med. 2014, 34, 100–105. [Google Scholar] [CrossRef]
- Shen, L.; Cong, W.J.; Lin, X.; Hong, Y.L.; Hu, R.W.; Feng, Y.; Xu, D.S.; Ruan, K.F. Characterization using LC/MS of the absorption compounds and metabolites in rat plasma after oral administration of a single or mixed decoction of Shaoyao and Gancao. Chem. Pharm. Bull. 2012, 60, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J. Progress and future developing of the serum pharmacochemistry of traditional Chinese medicine. China J. Chin. Mater. Med. 2006, 31, 789–792. [Google Scholar]
- Yang, X.; Peng, W.B.; Yue, X.Q. Syndrome differentiation and treatment of Taiyang disease in Shanghan Lun. J. Chin. Integr. Med. 2009, 7, 171–174. [Google Scholar] [CrossRef]
- Zhang, B.G.; Liu, Q.F. Clinic utilization of Guizhi decoction in modern times. China J. Chin. Mater. Med. 2007, 32, 757–760. [Google Scholar]
- Katakai, M.; Tani, T. A pair of crude drugs used in Shang-Han-Lun, especially the ways of using roasted licorice. Jpn. J. Hist. Pharm. 2003, 38, 151–160. [Google Scholar]
- Zhang, B.G.; Liang, X.X.; Liu, Q.F. Pharmacodynamics research of Guizhi decoction in modern times. China J. Chin. Mater. Med. 2007, 32, 557–561. [Google Scholar]
- Li, X.; Jiang, Y.H.; Jiang, P. Effect of Guizhi decoction on heart rate variability and regulation of cardiac autonomic nervous imbalance in diabetes mellitus rats. Chin. J. Integr. Med. 2014, 20, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, L.; Leng, J.; Chen, P.; Fan, X.; Cheng, Y. Fragment ion diagnostic strategies for the comprehensive identification of chemical profile of Gui-Zhi-Tang by integrating high-resolution MS, multiple-stage MS and UV information. J. Pharm. Biomed. Anal. 2014, 98, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Liang, H.; Cai, S.Q.; Zhao, Y.-Y. Studies on the chemical constituents of the active fraction of GuiZhi decoction. J. Chin. Pharm. Sci. 2007, 16, 24–26. [Google Scholar]
- Iwabu, J.; Watanabe, J.; Hirakura, K.; Ozaki, Y.; Hanazaki, K. Profiling of the compounds absorbed in human plasma and urine after oral administration of a traditional Japanese (Kampo) medicine, Daikenchuto. Drug Metab. Dispos. 2010, 38, 2040–2048. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Sugiyama, Y. Role of metabolic enzymes and efflux transporters in the absorption of drugs from the small intestine. Eur. J. Pharm. Sci. 2000, 12, 3–12. [Google Scholar] [CrossRef]
- Moscovitz, J.E.; Aleksunes, L.M. Establishment of metabolism and transport pathways in the rodent and human fetal liver. Int. J. Mol. Sci. 2013, 14, 23801–23827. [Google Scholar] [CrossRef] [PubMed]
- Herbert, M. Forensic relevance of glucuronidation in phase-II-metabolism of alcohols and drugs. Leg. Med. 2009, 11, S22–S26. [Google Scholar]
- Kampe, T.; König, A.; Schroeder, H.; Hengstler, J.G.; Niemeyer, C.M. Modular microfluidic system for emulation human phase I/phase II metabolism. Anal. Chem. 2014, 86, 3068–3074. [Google Scholar] [CrossRef] [PubMed]
- Mullen, W.; Boitier, A.; Stewart, A.J.; Crozier, A. Flavonoid metabolites in human plasma and urine after the consumption of red onions: Analysis by liquid chromatography with photodiode array and full scan tandem mass spectrometric detection. J. Chromatogr. A 2004, 1058, 163–168. [Google Scholar] [CrossRef]
- Miners, J.O.; Smith, P.A.; Sorich, M.J.; McKinnon, R.A.; Mackenzie, P.I. Predicting human drug glucuronidation parameters: Application of in vitro and in silico modeling approaches. Annu. Rev. Pharmacol. Toxicol. 2004, 44, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.J.; Qiu, S.X.; Wufuer, A.; Shum, L. Simultaneous determination of glycyrrhizin, a marker component in radix Glycyrrhizae, and its major metabolite glycyrrhetic acid in human plasma by LC–MS/MS. J. Chromatogr. B 2005, 814, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.R.; Ma, Y.M.; Ma, W. Pharmacokinetics and bioavailability of cinnamic acid after oral administration of Ramulus Cinnamomi in rats. Eur. J. Drug Metab. Pharmacokinet. 2009, 34, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Xu, F.; Zhang, Y.Z.; Huang, S.; Zang, X.Y.; Zhao, X.; Zhang, L.; Shang, M.Y.; Yang, D.H.; Wang, X.; Cai, S.Q. The profiling and identification of the absorbed constituents and metabolites of Paeoniae Radix Rubra decoction in rat plasma and urine by the HPLC-DAD-ESI-IT-TOF-MSn technique: A novel strategy for the systematic screening and identification of absorbed constituents and metabolites from traditional Chinese medicines. J. Pharm. Biomed. Anal. 2013, 83, 108–121. [Google Scholar] [PubMed]
- Yan, Z.; Chen, Y.; Li, T.; Zhang, J.; Yang, X. Identification of metabolites of Si-Ni-San, a traditional Chinese medicine formula, in rat plasma and urine using liquid chromatography/diode array detection/triple–quadrupole spectrometry. J. Chromatogr. B 2012, 885, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Jack, J.W.; Rendan, B.W.; Ferruzzi, M.G.; Reuhs, B.L.; Cooper, B.R.; Harwood, J.S.; Shulaev, V.; Pasinetti, G.; Dixon, R.A. Synthesis and Quantitative Analysis of Plasma-Targeted Metabolites of Catechin and Epicatechin. J. Agric. Food Chem. 2015, 63, 2233–2240. [Google Scholar]
- Shali, N.A.; Curtis, G.M.; Powell, A.B. Sulfation of the flavonoids quercetin and catechin by rat liver. Xenobiotica 1991, 21, 881–893. [Google Scholar] [CrossRef] [PubMed]
- Gonthier, M.P.; Donovan, J.L.; Texier, O.; Felgines, C.; Remesy, C.; Scalbert, A. Metabolism of dietary procyanidins in rats. Free Radic. Biol. Med. 2003, 35, 837–844. [Google Scholar] [CrossRef]
- Tourino, S.; Perez-Jimenez, J.; Mateos-Martin, M.L.; Fuguet, E.; Vinardell, M.P.; Cascante, M.; Torres, J.L. Metabolites in contact with rat digestive tract after ingestion of a phenolic-rich dietary fiber matrix. J. Agric. Food Chem. 2011, 59, 5955–5963. [Google Scholar] [CrossRef] [PubMed]
- Urpi-Sarda, M.; Garrido, I.; Monagas, M.; Gómez-Cordovés, C.; Medina-Remón, A.; Andres-Lacueva, C.; Bartolomé, B. Profile of plasma and urine metabolites after the intake of almond [Prunus dulcis (Mill.) DA Webb] polyphenols in humans. J. Agric. Food Chem. 2009, 57, 10134–10142. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Xu, F.; Zhang, Y.Z.; Zang, X.-Y.; Wang, D.; Shang, M.-Y.; Wang, X.; Chui, D.-H.; Cai, S.-Q. The profiling and identification of the metabolites of (+)-catechin and study on their distribution in rats by HPLC-DAD-ESI-IT-TOF-MSn technique. Biomed. Chromatogr. 2014, 28, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, T.; Inaba, A.; Ohmori, M.; Endo, T.; Kubo, S.; Ohsawa, K. Urinary metabolites of gallic acid in rats and their radical-scavenging effects on 1, 1-diphenyl-2-picrylhydrazyl radical. J. Nat. Prod. 2000, 63, 1444–1446. [Google Scholar] [CrossRef] [PubMed]
- Bredsdorff, L.; Nielsen, I.L.F.; Rasmussen, S.E.; Cornett, C.; Barron, D.; Bouisset, F.; Offord, E.; Williamson, G. Absorption, conjugation and excretion of the flavanones, naringenin and hesperetin from α-rhamnosidase-treated orange juice in human subjects. Br. J. Nutr. 2010, 103, 1602–1609. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.; Liu, M.; Dong, X.; Wu, S.; Fan, L.; Qiao, Y.; Chai, Y.; Wu, H. A strategy for rapid analysis of xenobiotic metabolome of Sini decoction in vivo using ultra-performance liquid chromatography-electrospray ionization quadrupole-time-of-flight mass spectrometry combined with pattern recognition approach. J. Pharm. Biomed. Anal. 2014, 96, 187–196. [Google Scholar] [CrossRef] [PubMed]






| No. | tR (min) | Identification | Formula | Negative Ion (m/z) | Positive Ion (m/z) | Source | ||
|---|---|---|---|---|---|---|---|---|
| Quasi-Molecular (ppm) | MS/MS (m/z) | Quasi-Molecular (ppm) | MS/MS (m/z) | |||||
| A1 | 0.945 | cAMP | C10H12N5O6P | 328.0459 | 134.0463 | 330.0590 | 136.0618 | ZJ |
| A2 | 1.194 | Gallic acid | C7H6O5 | 169.0133 | 125.0239 | - | - | P |
| A3 | 6.804 | Epicatechin | C15H14O6 | 289.0728 | 245.0824, 203.0730, 125.0824,109.0306 | - | - | P |
| A4 | 7.035 | Oxypaeoniflorin isomer I | C23H28O12 | - | - | 497.1649 | 197.0811, 179.0701, 151.0751, 121.0285 | P |
| A5 | 7.685 | Catechin | C15H14O6 | 289.0718 | 245.0827, 203.0720, 151.0398 | 291.0858 | 207.0669, 179.0705, 147.0432, 139.0388 | P |
| A6 | 7.762 | Oxypaeoniflorin | C23H28O12 | 495.1508 | 465.1381, 333.0985, 255.0670, 177.0554 | - | - | P |
| A7 | 10.199 | Albiflorin | C23H28O11 | 525.1630 [M+HCOO−H]− | 121.0294 | 481.1706 | 197.0811 | P |
| A8 | 10.391 | Liquiritigenin-7-O-glucoside-4’-O-apiosyl-O-glucoside | C32H40O18 | 711.2131 | 549.1615 | - | - | G |
| A9 | 10.856 | Paeoniflorin | C23H28O11 | 525.1619 [M+HCOO−H]− | 449.1453, 327.1082, 165.0541, 121.0291 | - | - | P |
| A10 | 11.140 | Mudanpioside I | C23H28O11 | 479.1664 | 121.0290 | 481.1703 | 179.0709 | P |
| A11 | 11.405 | Oxypaeoniflorin isomer II | C23H28O12 | 495.1513 | 137.0243 | - | - | P |
| A12 | 12.305 | Naringenin-7-O-glucoside | C21H22O10 | 433.1154 | 271.0612, 151.0034, 119.0503 | - | - | G |
| A13 | 12.423 | Liquiritigenin-7-O-glucoside-4’-O-apiosyl-O-glucoside isomer I | C32H40O18 | 711.2131 | 549.1615 | - | - | G |
| A14 | 12.704 | 4-Shogoal | C15H20O3 | - | - | 249.1500 | 177.0920, 137.0604 | ZR |
| A15 | 12.710 | Liquiritin | C21H22O9 | 417.1189 | 255.0654 | - | - | G |
| A16 | 12.810 | Liquiritin-apioside | C26H30O13 | 549.1589 | 255.0662 | - | - | G |
| A17 | 12.825 | Rutin | C27H30O16 | 609.1451 | 301.1452 | 611.1645 | 303.0517 | ZJ |
| A18 | 13.245 | Galloylpaeoniflorin | C30H32O15 | 631.1660 | 465.1385, 313.0556, 271.0454 | 633.1814 | 315.0705, 179.0700, 153.0179, 127.0390 | P |
| A19 | 13.404 | Kaempferol-3-O-rutinoside | C27H30O15 | 593.1537 | 285.0394 | 595.1698 | 287.0563 | ZJ |
| A20 | 14.277 | Galloylpaeoniflorin isomer I | C30H32O15 | 631.1667 | 477.0935, 271.0604, 121.0290 | - | - | P |
| A21 | 14.351 | Naringenin-5-O-glucoside | C21H22O10 | 433.1154 | 271.0609, 151.0032 | - | - | G |
| A22 | 14.706 | Paeoniflorin isomer I | C23H28O11 | 525.1630 [M+HCOO−H]− | 121.0291 | 481.1709 | 197.0809, 105.0339 | P |
| A23 | 15.056 | Lactiflorin | C23H26O10 | 507.1519 [M+HCOO−H]− | 461.1430, 283.0631, 177.0555, 121.0292 | - | - | P |
| A24 | 15.400 | Lsoliquiritin-apioside | C26H30O13 | 549.1620 | 255.0662 | - | - | G |
| A25 | 15.538 | Ononin | C22H22O9 | 475.1256 [M+HCOO−H]− | 267.0651, 252.0416 | 431.1336 | 269.0809 | G |
| A26 | 15.714 | Lsoliquiritin | C21H22O9 | 417.1150 | 225.0659, 135.0086, 119.0500 | - | - | G |
| A27 | 15.813 | 5-hydroxylliquiritin | C21H22O10 | 433.1146 | 271.0607 | - | - | G |
| A28 | 16.544 | Liquiritigenin | C15H12O4 | 255.0671 | 199.0508, 135.0094 | - | - | G |
| A29 | 17.605 | 10-Gingerdiol | C21H36O4 | - | - | 376.1810 | 259.1711, 137.0609 | ZR |
| A30 | 18.112 | Licorice saponin G2 | C42H62O17 | 837.3917 | 351.0576 | 839.4062 | 663.3724, 469.3306 | G |
| A31 | 18.125 | 8-Gingerdione | C19H28O4 | - | - | 321.2217 | 177.0923, 137.0604 | ZR |
| A32 | 18.638 | Pentagalloylglucose | C41H32O26 | 939.1109 | - | - | - | P |
| A33 | 18.679 | 10-Gingerol | C21H34O4 | - | - | 373.1668 [M+Na]+ | 351.2658, 207.1017, 177.0557, 137.0605 | ZR |
| A34 | 18.781 | Benzoylpaeoniflorin | C41H32O26 | 629.1876, [M+HCOO−H]− | 553.1716, 431.1349, 165.0558, 121.0291 | - | - | P |
| A35 | 18.977 | Apioglycyrrhizin | C42H62O16 | 821.3949 | - | - | - | G |
| A36 | 19.334 | Benzoylpaeoniflorin isomer | C30H32O12 | 629.1881 [M+HCOO−H]− | 121.0293 | 607.1770 [M+Na]+ | 319.1183, 267.0860, 197.0808, 151.0340 | P |
| A37 | 19.542 | Licorice saponin A3 | C48H72O21 | 983.4484 | 821.3958, 351.0559 | 985.4642 | 809.4310, 615.3888, 453.3359 | G |
| A38 | 20.278 | Acetoxyglycyrrhizin acid | C44H64O18 | - | - | 881.4165 | 705.3835, 511.3421 | G |
| A39 | 20.503 | Licorice saponin G2 isomer I | C42H62O17 | 837.3904 | - | 839.4058 | 663.3730, 469.3310 | G |
| A40 | 21.153 | 8-Gingerdiol | C19H32O4 | - | - | 326.1860 | 137.0605 | ZR |
| A41 | 21.354 | Formononetin | C16H12O4 | 267.0661 | 252.0415, 223.5401 | 269.0806 | 254.0850, 225.0557 | G |
| A42 | 21.510 | Isoliquiritigenin | C15H12O4 | 255.0671 | 135.0094, 119.0498 | - | - | G |
| A43 | 21.767 | Licorice saponin E2 | C42H60O16 | 819.3816 | 351.0548 | - | - | G |
| A44 | 21.960 | Licorice saponin G2 isomer II | C42H62O17 | 837.3907 | - | 839.4058 | 645.3617, 469.3312, 451.3203 | G |
| A45 | 22.636 | Licorice saponin G2 isomer III | C42H62O17 | - | - | 839.4060 | 487.3406, 469.3307, 451.3193 | G |
| A46 | 22.636 | 22β-Acetoxyllicoricesaponin C2 | C44H64O17 | 863.4066 | - | 865.4217 | 495.3466 | G |
| A47 | 23.051 | Licorice saponin G2 isomer IV | C44H62O17 | 837.3918 | - | 839.4069 | 487.3415, 469.3307, 451.3215 | G |
| A48 | 23.182 | 6-Paradol | C17H26O3 | - | - | 279.1969 | 177.0940, 163.0755, 145.0661, 137.0601 | ZR |
| A49 | 23.182 | 8-Dehydrogingerdione | C19H26O4 | - | - | 319.1886 | 177.0916, 163.0751, 145.0659, 137.0604 | ZR |
| A50 | 23.208 | 6-Gingerdiol | C17H28O4 | - | - | 297.2057 | 177.0923, 163.0752, 137.0595 | ZR |
| A51 | 23.416 | Glycyrrhizic acid | C42H62O16 | 821.3952 | - | 823.4109 | 647.3780, 453.3368 | G |
| A52 | 23.935 | Licorice saponin G2 isomer IV | C42H62O17 | - | - | 839.4046 | 487.3423, 469.3307, 451.3203 | G |
| A53 | 23.961 | 6-Gingerol | C17H26O4 | - | - | 295.1916 | 163.0757, 137.0605 | ZR |
| A54 | 24.117 | Uralenol | C20H18O7 | 369.1353 | 229.0864, 139.0395 | 371.1493 | 315.0874, 175.0398 | G |
| A55 | 24.403 | 6-Shogaol | C17H24O3 | - | - | 277.1790 | 177.0675, 145.0647, 137.0598 | ZR |
| A56 | 24.482 | LS-K2 | C42H62O16 | 821.3942 | - | 823.4411 | 647.3785, 453.3361 | G |
| A57 | 24.664 | Apioglycyrrhizin | C42H62O16 | 821.3949 | - | 823.4111 | 647.3780, 453.3362 | G |
| A58 | 24.755 | 6-Gingerdione | C17H24O4 | - | - | 293.1773 | 177.0557, 145.0295, 137.0604 | ZR |
| A59 | 25.756 | LS-J2 | C42H64O16 | 823.4130 | 351.0554 | 825.4303 | 453.3377 | G |
| A60 | 26.458 | LS-C2 | C42H62O15 | 805.4300 | 351.0565 | 825.4300 [M+H2O+H]+ | 437.3435, 353.0731 | G |
| A61 | 26.640 | 10-Gingerdione | C21H32O4 | - | - | 349.1784 | 177.0917, 137.0591 | ZR |
| A62 | 26.718 | Glycycoumarin | C21H20O6 | 367.1184 | 309.0402, 297.0405 | 369.1335 | 313.0725, 285.0769 | G |
| A63 | 27.472 | 7-Shogaol | C18H26O3 | - | - | 291.1974 | 177.0918, 137.0606 | ZR |
| A64 | 28.278 | Licoricone | C22H22O6 | 381.1340 | 351.0869, 323.0593 | 383.1508 | 327.0872, 299.0934 | G |
| A65 | 28.954 | 10-Dehydrogingerdione | C21H30O4 | - | - | 347.2188 | 177.0835, 137.0611 | ZR |
| A66 | 29.344 | Isoglycyrol | C21H18O6 | 365.1039 | 307.0247, 295.0247 | 367.1265 | 339.1254, 311.0562 | G |
| A67 | 30.020 | 8-Shogaol | C19H28O3 | - | - | 305.2118 | 177.0921, 137.0604 | ZR |
| A68 | 30.020 | 8-Gingerol | C19H30O4 | - | - | 345.2042 | 177.0908, 137.0596 | ZR |
| A69 | 33.791 | 10-Shogaol | C21H32O3 | - | - | 333.2431 | 177.0910, 137.0595 | ZR |
| A70 | 34.519 | Cinnamic acid | C9H8O2 | - | - | 149.0235 | 121.0283 | C |
| A71 | 38.121 | Oleanolic acid | C30H38O3 | 455.3533 | - | - | - | ZJ |
| No. | tR (min) | Identification | Formula | Urine | Plasma | Negative Ion (m/z) | Possible Original Compound | Source | |
|---|---|---|---|---|---|---|---|---|---|
| Quasi-Molecular (ppm) | MS/MS (m/z) | ||||||||
| M1 | 4.494 | Catechin-O-glucuronide | C21H22O12 | + | - | 465.0785 | 289.0622, 245.0724 | (Epi) catechin-related | P |
| M2 | 9.026 | 3’-O-Methyl (epi)catechin 5-O-glucuronide | C21H24O12 | + | - | 479.0965 | 303.0976, 175.0176 | (Epi) catechin-related | P |
| M3 | 10.220 | 3’-O-Methyl (epi)catechin 7- or 4’-O-glucuronide | C21H24O12 | - | + | 479.0948 | 303.0876, 175.0242 | (Epi) catechin-related | P |
| M4 | 8.861 | 5-(3,4-Dihydroxyphenyl)-γ-valerolactone glucuronide | C17H20O10 | + | - | 383.0757 | 207.0587, 163.0691 | (Epi) catechin-related | P |
| M5 | 5.949 | 5-(3,4-Dihydroxyphenyl)-γ-valerolactone sulfate | C11H12O7S | + | - | 287.0027 | 207.0580, 163.0685 | (Epi) catechin-related | P |
| M6 | 17.003 | 5-(3-Methoxyl-4-hydroxyphenyl)-valerolactone glucuronide | C18H22O10 | + | - | 397.0907 | 221.0719 | (Epi) catechin-related | P |
| M7 | 19.489 | 5-(3-Methoxyl-4-hydroxyphenyl)-valerolactone sulfate | C12H14O7S | + | - | 301.0187 | 221.0739, 206.0503 | (Epi) catechin-related | P |
| M8 | 9.305 | 5-(3-Hydroxyphenyl)-γ-valerolactone sulfate | C11H12O6S | + | - | 271.0092 | 191.0624 | (Epi) catechin-related | P |
| M9 | 4.276 | 4-Hydroxy phenylpropionic acid sulfate | C9H10O6S | + | - | 244.9955 | 165.0596 | (Epi) catechin-related | P |
| M10 | 9.549 | 3-Hydroxy phenylpropionic acid sulfate | C9H10O6S | + | - | 245.0320 | 165.0846 | (Epi) catechin-related | P |
| M11 | 8.547 | 3,4-Dihydroxy phenylpropionic acid sulfate | C9H10O7S | + | - | 260.9888 | 181.0409, 166.0193 | (Epi) catechin-related | P |
| M12 | 2.022 | 3-Hydroxy-4-methoxy-phenylpropionic acid sulfate | C10H12O7S | + | - | 275.0040 | 195.0717 | (Epi) catechin-related | P |
| M13 | 5.027 | 3-Methoxy-4-hydroxy-phenylpropionic acid sulfate | C10H12O7S | + | + | 275.0042 | 195.0504 | (Epi) catechin-related | P |
| M14 | 4.511 | m-Coumaric acid sulfate | C9H8O6S | + | - | 242.9793 | 163.0324 | (Epi) catechin-related | P |
| M15 | 14.077 | p-Coumaric acid sulfate | C9H8O6S | + | - | 243.0173 | 163.0694 | (Epi) catechin-related | P |
| M16 | 10.666 | Ferulic acid sulfate | C10H10O7S | + | + | 273.0259 | 193.0845 | (Epi) catechin-related | P |
| M17 | 9.057 | 3-Hydroxy-4-methoxyphenylacetic acid sulfate | C9H10O7S | + | - | 261.0254 | 181.0718 | (Epi) catechin-related | P |
| M18 | 1.661 | 3,4-Dihydroxy phenylacetic acid sulfate | C8H8O7S | + | - | 247.0111 | 167.0575 | (Epi) catechin-related | P |
| M19 | 1.713 | Protocatechuic acid-3- or -4-O-sulfate | C7H6O7S | + | - | 232.9967 | 153.0423 | (Epi) catechin-related | P |
| M20 | 20.086 | Benzoyl glucuronide | C13H14O8 | + | - | 297.0939 | 121.0658 | (Epi) catechin-related | P |
| M21 | 9.92 | 3-Hydroxyhipuric acid | C9H9NO4 | + | - | 194.0298 | 150.0462 | (Epi) catechin-related | P |
| M22 | 13.393 | Gallic acid glucuronide | C13H14O11 | + | + | 345.1354 | 169.1232 | Gallic acid-related | P |
| M23 | 17.521 | 4-O-Methylgallic acid | C8H8O5 | + | - | 183.0887 | 168.0197, 139.0268 | Gallic acid-related | P |
| M24 | 14.378 | 4-O-Methylgallic acid glucuronide | C14H16O11 | + | - | 359.1115 | 191.0628, 183.0944 | Gallic acid-related | P |
| M25 | 1.202 | 2-Deoxy-pyrogallol-1-O-sulfate | C6H6O5S | + | - | 188.9718 | 125.0010, 109.0087 | Gallic acid-related | P |
| M26 | 0.845 | Pyrogallol-2-O-sulfate | C6H6O6S | + | - | 204.9665 | 125.0126 | Gallic acid-related | P |
| M27 | 1.254 | Pyrogallol-1-O-sulfate | C6H6O6S | + | - | 204.9665 | 124.9982 | Gallic acid-related | P |
| M28 | 30.462 | 2-O-Methylpyrogallol glucuronide | C13H16O9 | + | - | 315.2336 | 171.1027, 139.1119 | Gallic acid-related | P |
| M29 | 0.888 | 2-O-Methylpyrogallol sulfate | C7H8O6S | + | - | 218.9790 | 139.0419, 124.0168 | Gallic acid-related | P |
| M30 | 1.228 | 1-O-Methylpyrogallol-3-O-sulfate | C7H8O6S | + | - | 218.9809 | 139.0277 | Gallic acid-related | P |
| M31 | 2.021 | 1-O-Methylpyrogallol-2-O-sulfate | C7H8O6S | + | - | 218.9809 | 139.0273 | Gallic acid-related | P |
| M32 | 17.146 | Paeonimetabolin I glucuronide isomer I or II | C16H22O10 | + | - | 373.1311 | 197.1181 | Paeoniflorin-related | P |
| M33 | 8.939 | C10H14O3 sulfate | C10H14O6S | + | - | 260.9913 | 181.0443 | Paeoniflorin-related | P |
| M34 | 14.952 | C10H18O2 glucuronide | C16H26O8 | + | - | 345.1354 | 169.1223 | Paeoniflorin-related | P |
| M35 | 19.135 | Naringenin | C15H12O5 | + | - | 271.0419 | 151.0032 | Naringenin-O-glucoside -related | G |
| M36 | 16.194 | Naringenin-O-glucuronide-O-sulfate | C21H20O14S | + | - | 527.0264 | 447.0951, 351.0192, 271.0628 | Naringenin-O-glucoside-related | G |
| M37 | 15.292 | Naringenin-4’-O-glucuronide | C21H20O11 | + | - | 447.1025 | 271.0887, 175.0160 | Naringenin-O-glucoside-related | G |
| M38 | 17.312 | Naringenin-7-O-glucuronide | C21H20O11 | + | - | 447.1025 | 271.0887, 175.0160, 150.9851 | Naringenin-O-glucoside-related | G |
| M39 | 13.133 | Naringenin-O-sulfate | C15H12O8S | + | - | 350.9982 | 271.0390, 150.9829 | Naringenin-O-glucoside-related | G |
| M40 | 16.690 | Liquiritigenin-O-glucuronide-O-sulfate | C21H20O13S | + | - | 511.0306 | 431.0900, 335.0135 | Liquiritigenin-related | G |
| M41 | 13.386 | 7,4’-Dihydroxyflavone-O-sulfate | C15H10O7S | + | - | 332.9885 | 253.0431 | Liquiritigenin-related | G |
| M42 | 13.619 | Liquiritigenin-4’-O-sulfate | C15H12O7S | + | - | 335.0018 | 255.0569, 134.9995 | Liquiritigenin-related | G |
| M43 | 14.230 | Liquiritigenin-7-O-sulfate | C15H12O7S | + | - | 335.0018 | 255.0658, 135.0088 | Liquiritigenin-related | G |
| M44 | 12.532 | Liquiritigenin-4’-O-glucuronide | C21H20O10 | + | - | 431.0754 | 255.0498, 113.0137 | Liquiritigenin-related | G |
| M45 | 12.924 | Liquiritigenin-7-O-glucuronide | C21H20O10 | + | - | 431.0754 | 255.0499, 113.0132 | Liquiritigenin-related | G |
| M46 | 29.631 | Isoliquiritigenin-O-sulfate | C15H12O7S | + | - | 335.2224 | 255.2111 | Isoliquiritigenin-related | G |
| M47 | 16.023 | Isoliquiritigenin-4’-O-glucuronide | C21H20O10 | + | + | 431.0754 | 255.0672, 113.0247 | Isoliquiritigenin-related | G |
| M48 | 16.441 | Isoliquiritigenin-7-O-glucuronide | C21H20O10 | + | - | 431.0754 | 255.0559, 113.0163 | Isoliquiritigenin-related | G |
| M42 | 13.619 | Liquiritigenin-4’-O-sulfate | C15H12O7S | + | - | 335.0018 | 255.0569, 134.9995 | Liquiritigenin-related | G |
| M43 | 14.230 | Liquiritigenin-7-O-sulfate | C15H12O7S | + | - | 335.0018 | 255.0658, 135.0088 | Liquiritigenin-related | G |
| M44 | 12.532 | Liquiritigenin-4’-O-glucuronide | C21H20O10 | + | - | 431.0754 | 255.0498, 113.0137 | Liquiritigenin-related | G |
| M45 | 12.924 | Liquiritigenin-7-O-glucuronide | C21H20O10 | + | - | 431.0754 | 255.0499, 113.0132 | Liquiritigenin-related | G |
| M46 | 29.631 | Isoliquiritigenin-O-sulfate | C15H12O7S | + | - | 335.2224 | 255.2111 | Isoliquiritigenin-related | G |
| M47 | 16.023 | Isoliquiritigenin-4’-O-glucuronide | C21H20O10 | + | + | 431.0754 | 255.0672, 113.0247 | Isoliquiritigenin-related | G |
| M48 | 16.441 | Isoliquiritigenin-7-O-glucuronide | C21H20O10 | + | - | 431.0754 | 255.0559, 113.0163 | Isoliquiritigenin-related | G |
| M49 | 18.463 | Fomononetin-O-sulfate | C16H12O7S | + | - | 347.0012 | 267.0637 | Fomononetin-related | G |
| No. | tR (min) | Identification | Formula | Urine | Plasma | Positive Ion (m/z) | Possible Original Compound | Source | |
|---|---|---|---|---|---|---|---|---|---|
| Quasi-molecular (ppm) | MS/MS (m/z) | ||||||||
| M50 | 34.133 | Glycyrrhetinic acid | C30H46O4 | + | + | 471.3544 | 317.2164 | Glycyrrhizin-related | G |
| M51 | 23.150 | 6-Gingerol-O-glucuronide | C23H34O10 | + | - | 471.1015 | 295.0564 | 6-Gingerol-related | ZR |
| M52 | 0.524 | Hippuric acid | C9H8NO3 | + | + | 178.1015 | - | Cinnammic acid-related | C |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiang, H.; Zhang, L.; Song, J.; Fan, B.; Nie, Y.; Bai, D.; Lei, H. The Profiling and Identification of the Absorbed Constituents and Metabolites of Guizhi Decoction in Rat Plasma and Urine by Rapid Resolution Liquid Chromatography Combined with Quadrupole-Time-of-Flight Mass Spectrometry. Int. J. Mol. Sci. 2016, 17, 1409. https://doi.org/10.3390/ijms17091409
Xiang H, Zhang L, Song J, Fan B, Nie Y, Bai D, Lei H. The Profiling and Identification of the Absorbed Constituents and Metabolites of Guizhi Decoction in Rat Plasma and Urine by Rapid Resolution Liquid Chromatography Combined with Quadrupole-Time-of-Flight Mass Spectrometry. International Journal of Molecular Sciences. 2016; 17(9):1409. https://doi.org/10.3390/ijms17091409
Chicago/Turabian StyleXiang, Hongjun, Lishi Zhang, Jiannan Song, Bin Fan, Yinglan Nie, Dong Bai, and Haimin Lei. 2016. "The Profiling and Identification of the Absorbed Constituents and Metabolites of Guizhi Decoction in Rat Plasma and Urine by Rapid Resolution Liquid Chromatography Combined with Quadrupole-Time-of-Flight Mass Spectrometry" International Journal of Molecular Sciences 17, no. 9: 1409. https://doi.org/10.3390/ijms17091409
APA StyleXiang, H., Zhang, L., Song, J., Fan, B., Nie, Y., Bai, D., & Lei, H. (2016). The Profiling and Identification of the Absorbed Constituents and Metabolites of Guizhi Decoction in Rat Plasma and Urine by Rapid Resolution Liquid Chromatography Combined with Quadrupole-Time-of-Flight Mass Spectrometry. International Journal of Molecular Sciences, 17(9), 1409. https://doi.org/10.3390/ijms17091409