Analysis of Large Seeds from Three Different Medicago truncatula Ecotypes Reveals a Potential Role of Hormonal Balance in Final Size Determination of Legume Grains
Abstract
:1. Introduction
2. Results
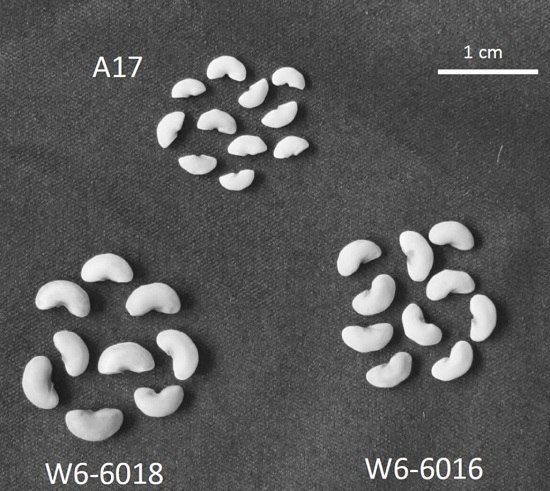
2.1. Identification of Medicago Accessions with Larger Seeds and Different Growth Patterns
2.2. Embryo Development Was Delayed in the Large Seed Accessions
2.3. The Increase of Mature Seed Size Was Due to Increase in Cell Division but Not Cell Expansion
2.4. Change in ABA to IAA Ratio Correlates with the Variation of Seed Size
3. Discussion
3.1. Increase of Seed Size as a Result of Increase of Cell Division during Embryogenesis
3.2. Phytohormones as Regulators of Cell Division
4. Experimental Section
4.1. Plant Material and Growth Conditions
4.2. Fresh and Dry Weight Measurement
4.3. Clearing of Seeds for DIC Imaging
4.4. Counting Total Number of Cells in Mature Seeds
4.5. Counting Number of Cells in a Given Area (Cell Size)
4.6. Phytohormone Measurement
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Moles, A.T.; Ackerly, D.D.; Webb, C.O.; Tweddle, J.C.; Dickie, J.B.; Westoby, M. A brief history of seed size. Science 2005, 307, 576–580. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Li, Y. Ubiquitin-mediated control of seed size in plants. Front. Plant Sci. 2014, 5, 332. [Google Scholar] [CrossRef] [PubMed]
- Sreenivasulu, N.; Wobus, U. Seed-development programs: A systems biology-based comparison between dicots and monocots. Annu. Rev. Plant Biol. 2013, 64, 189–217. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, V. Control of seed size in plants. Proc. Natl. Acad. Sci. USA 2005, 102, 17887–17888. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, R.B.; de Paiva, G.; Yadegari, R. Plant embryogenesis: Zygote to seed. Science 1994, 266, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Venglat, P.; Xiang, D.; Wang, E.; Datla, R. Genomics of seed development: Challenges and opportunities for genetic improvement of seed traits in crop plants. Biocatal. Agric. Biotechnol. 2014, 3, 24–30. [Google Scholar] [CrossRef]
- Jofuku, K.D.; Omidyar, P.K.; Gee, Z.; Okamuro, J.K. Control of seed mass and seed yield by the floral homeotic gene apetala2. Proc. Natl. Acad. Sci. USA 2005, 102, 3117–3122. [Google Scholar] [CrossRef] [PubMed]
- Ohto, M.A.; Fischer, R.L.; Goldberg, R.B.; Nakamura, K.; Harada, J.J. Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. USA 2005, 102, 3123–3128. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Dennis, E.S.; Berger, F.; Peacock, W.J.; Chaudhury, A. Miniseed3 (MINI3), a wrky family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) kinase gene, are regulators of seed size in arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 17531–17536. [Google Scholar] [CrossRef] [PubMed]
- Adamski, N.M.; Anastasiou, E.; Eriksson, S.; O’Neill, C.M.; Lenhard, M. Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 20115–20120. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Sun, S.; Yao, J.; Wang, C.; Yu, S.; Xu, C.; Li, X.; Zhang, Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 19579–19584. [Google Scholar] [CrossRef] [PubMed]
- Disch, S.; Anastasiou, E.; Sharma, V.K.; Laux, T.; Fletcher, J.C.; Lenhard, M. The E3 ubiquitin ligase big brother controls arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 2006, 16, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Li, N.; Dumenil, J.; Li, J.; Kamenski, A.; Bevan, M.W.; Gao, F.; Li, Y. The ubiquitin receptor da1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in arabidopsis. Plant Cell 2013, 25, 3347–3359. [Google Scholar] [CrossRef] [PubMed]
- Young, N.D.; Debelle, F.; Oldroyd, G.E.; Geurts, R.; Cannon, S.B.; Udvardi, M.K.; Benedito, V.A.; Mayer, K.F.; Gouzy, J.; Schoof, H.; et al. The medicago genome provides insight into the evolution of rhizobial symbioses. Nature 2011, 480, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Chabaud, M.; de Carvalho-Niebel, F.; Barker, D.G. Efficient transformation of Medicago truncatula cv. Jemalong using the hypervirulent agrobacterium tumefaciens strain agl1. Plant Cell Rep. 2003, 22, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Tadege, M.; Wen, J.; He, J.; Tu, H.; Kwak, Y.; Eschstruth, A.; Cayrel, A.; Endre, G.; Zhao, P.X.; Chabaud, M.; et al. Large-scale insertional mutagenesis using the tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008, 54, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Le Signor, C.; Savois, V.; Aubert, G.; Verdier, J.; Nicolas, M.; Pagny, G.; Moussy, F.; Sanchez, M.; Baker, D.; Clarke, J.; et al. Optimizing tilling populations for reverse genetics in Medicago truncatula. Plant Biotechnol. J. 2009, 7, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Rogers, C.; Wen, J.; Chen, R.; Oldroyd, G. Deletion-based reverse genetics in Medicago truncatula. Plant Physiol. 2009, 151, 1077–1086. [Google Scholar] [CrossRef] [PubMed]
- Benedito, V.A.; Torres-Jerez, I.; Murray, J.D.; Andriankaja, A.; Allen, S.; Kakar, K.; Wandrey, M.; Verdier, J.; Zuber, H.; Ott, T.; et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008, 55, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Young, N.D.; Udvardi, M. Translating Medicago truncatula genomics to crop legumes. Curr. Opin. Plant Biol. 2009, 12, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Verdier, J.; Dessaint, F.; Schneider, C.; Abirached-Darmency, M. A combined histology and transcriptome analysis unravels novel questions on Medicago truncatula seed coat. J. Exp. Bot. 2013, 64, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Noguero, M.; Le Signor, C.; Vernoud, V.; Bandyopadhyay, K.; Sanchez, M.; Fu, C.; Torres-Jerez, I.; Wen, J.; Mysore, K.S.; Gallardo, K.; et al. Dash transcription factor impacts Medicago truncatula seed size by its action on embryo morphogenesis and auxin homeostasis. Plant J. 2015, 81, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, K.; Firnhaber, C.; Zuber, H.; Hericher, D.; Belghazi, M.; Henry, C.; Kuster, H.; Thompson, R. A combined proteome and transcriptome analysis of developing Medicago truncatula seeds: Evidence for metabolic specialization of maternal and filial tissues. Mol. Cell. Proteom. 2007, 6, 2165–2179. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, U.; Xu, W.; Liu, A. Transcriptome profiling identifies aba mediated regulatory changes towards storage filling in developing seeds of castor bean (Ricinus communis L.). Cell Biosci. 2014, 4, 33. [Google Scholar] [CrossRef] [PubMed]
- Schussler, J.R.; Brenner, M.L.; Brun, W.A. Abscisic acid and its relationship to seed filling in soybeans. Plant Physiol. 1984, 76, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Chojecki, A.J.S.; Gale, M.D.; Bayliss, M.W. The number and sizes of starch granules in the wheat endosperm, and their association with grain weight. Ann. Bot. 1986, 58, 819–831. [Google Scholar]
- Chojecki, A.J.S.; Bayliss, M.W.; Gale, M.D. Cell production and DNA accumulation in the wheat endosperm, and their association with grain weight. Ann. Bot. 1986, 58, 809–817. [Google Scholar]
- Munier-Jolain, N.; Ney, B. Seed growth rate in grain legumes ii. Seed growth rate depends on cotyledon cell number. J. Exp. Bot. 1998, 49, 1971–1976. [Google Scholar] [CrossRef]
- Abirached-Darmency, M.; Abdel-gawwad, M.R.; Conejero, G.; Verdeil, J.L.; Thompson, R. In situ expression of two storage protein genes in relation to histo-differentiation at mid-embryogenesis in Medicago truncatula and Pisum sativum seeds. J. Exp. Bot. 2005, 56, 2019–2028. [Google Scholar] [CrossRef] [PubMed]
- Ohto, M.A.; Floyd, S.K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Effects of apetala2 on embryo, endosperm, and seed coat development determine seed size in arabidopsis. Sex Plant Reprod. 2009, 22, 277–289. [Google Scholar] [CrossRef] [PubMed]
- D’Erfurth, I.; Le Signor, C.; Aubert, G.; Sanchez, M.; Vernoud, V.; Darchy, B.; Lherminier, J.; Bourion, V.; Bouteiller, N.; Bendahmane, A.; et al. A role for an endosperm-localized subtilase in the control of seed size in legumes. New Phytol. 2012, 196, 738–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapman, E.J.; Estelle, M. Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 2009, 43, 265–285. [Google Scholar] [CrossRef] [PubMed]
- Perrot-Rechenmann, C. Cellular responses to auxin: Division versus expansion. Cold Spring Harb. Perspect. Biol. 2010, 2, a001446. [Google Scholar] [CrossRef] [PubMed]
- Moller, B.; Weijers, D. Auxin control of embryo patterning. Cold Spring Harb. Perspect. Biol. 2009, 1, a001545. [Google Scholar] [CrossRef] [PubMed]
- Schruff, M.C.; Spielman, M.; Tiwari, S.; Adams, S.; Fenby, N.; Scott, R.J. The auxin response factor 2 gene of arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 2006, 133, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Weber, H.; Borisjuk, L.; Wobus, U. Molecular physiology of legume seed development. Annu. Rev. Plant Biol. 2005, 56, 253–279. [Google Scholar] [CrossRef] [PubMed]
- Karssen, C.M.; Brinkhorst-van der Swan, D.L.; Breekland, A.E.; Koornneef, M. Induction of dormancy during seed development by endogenous abscisic acid: Studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 1983, 157, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Raz, V.; Bergervoet, J.H.; Koornneef, M. Sequential steps for developmental arrest in arabidopsis seeds. Development 2001, 128, 243–252. [Google Scholar] [PubMed]
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, S15–S45. [Google Scholar] [PubMed]
- Slater, S.M.; Yuan, H.Y.; Lulsdorf, M.M.; Vandenberg, A.; Zaharia, L.I.; Han, X.; Abrams, S.R. Comprehensive hormone profiling of the developing seeds of four grain legumes. Plant Cell. Rep. 2013, 32, 1939–1952. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Qi, Q.; Schorr, P.; Cutler, A.J.; Crosby, W.L.; Fowke, L.C. Ick1, a cyclin-dependent protein kinase inhibitor from arabidopsis thaliana interacts with both cdc2a and cycd3, and its expression is induced by abscisic acid. Plant J. 1998, 15, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Atif, R.M.; Boulisset, F.; Conreux, C.; Thompson, R.; Ochatt, S.J. In vitro auxin treatment promotes cell division and delays endoreduplication in developing seeds of the model legume species Medicago truncatula. Physiol. Plant 2013, 148, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Emery, R.J.; Ma, Q.; Atkins, C.A. The forms and sources of cytokinins in developing white lupine seeds and fruits. Plant Physiol. 2000, 123, 1593–1604. [Google Scholar] [CrossRef] [PubMed]
- Bewley, J.D. Seed germination and dormancy. Plant Cell 1997, 9, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Finch–Savage, W.E.; Leubner–Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.B.; Huang, H.Y.; Hu, Y.W.; Zhu, S.W.; Wang, Z.Y.; Lin, W.H. Brassinosteroid regulates seed size and shape in arabidopsis. Plant Physiol. 2013, 162, 1965–1977. [Google Scholar] [CrossRef] [PubMed]





© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bandyopadhyay, K.; Uluçay, O.; Şakiroğlu, M.; Udvardi, M.K.; Verdier, J. Analysis of Large Seeds from Three Different Medicago truncatula Ecotypes Reveals a Potential Role of Hormonal Balance in Final Size Determination of Legume Grains. Int. J. Mol. Sci. 2016, 17, 1472. https://doi.org/10.3390/ijms17091472
Bandyopadhyay K, Uluçay O, Şakiroğlu M, Udvardi MK, Verdier J. Analysis of Large Seeds from Three Different Medicago truncatula Ecotypes Reveals a Potential Role of Hormonal Balance in Final Size Determination of Legume Grains. International Journal of Molecular Sciences. 2016; 17(9):1472. https://doi.org/10.3390/ijms17091472
Chicago/Turabian StyleBandyopadhyay, Kaustav, Orhan Uluçay, Muhammet Şakiroğlu, Michael K. Udvardi, and Jerome Verdier. 2016. "Analysis of Large Seeds from Three Different Medicago truncatula Ecotypes Reveals a Potential Role of Hormonal Balance in Final Size Determination of Legume Grains" International Journal of Molecular Sciences 17, no. 9: 1472. https://doi.org/10.3390/ijms17091472
APA StyleBandyopadhyay, K., Uluçay, O., Şakiroğlu, M., Udvardi, M. K., & Verdier, J. (2016). Analysis of Large Seeds from Three Different Medicago truncatula Ecotypes Reveals a Potential Role of Hormonal Balance in Final Size Determination of Legume Grains. International Journal of Molecular Sciences, 17(9), 1472. https://doi.org/10.3390/ijms17091472