Unraveling Massive Crocins Transport and Accumulation through Proteome and Microscopy Tools during the Development of Saffron Stigma
Abstract
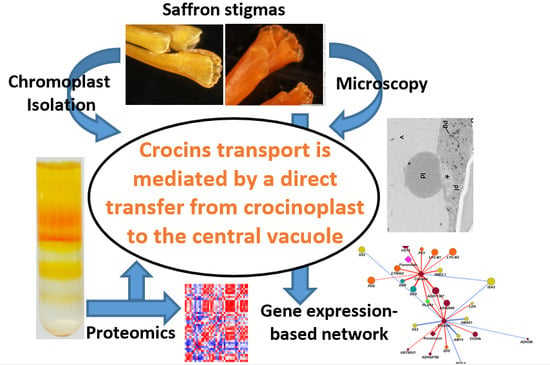
:1. Introduction
2. Results
2.1. Ultrastructural Characterization of Chromoplast Biogenesis in Saffron
2.2. Overall Proteomic Analysis
2.3. Proteins Involved in Isoprenoid Biosynthesis and Accumulation
2.4. Proteins of Interest in Sequestration and Related to Chromoplast Replication and Remodeling
2.5. Gene Expression Analysis of Genes Involved in Key Processes of Crocins Metabolism
2.6. Gene Expression Analyses of Starch Metabolism Genes: The Carbohydrate Supply for Apocarotenoids Biosynthesis
2.7. Correlation Analyses to Identify Candidate Genes Involved in Apocarotenoid Production
3. Discussion
4. Materials and Methods
4.1. Light and Transmission Electron Microscopy
4.2. Isolation and Purification of Saffron Chromoplasts from Fresh Stigmas
4.3. Extraction and Analysis of Crocins by HPLC-DAD, Marker-Enzyme Assays and Sequence Analysis
4.4. LC-MS/MS as an Analytical Method for the Identification of Chromoplast Proteins
4.5. Gene Expression by Quantitative Reverse Transcription-PCR (qRT-PCR)
4.6. Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Schmidt, M.; Betti, G.; Hensel, A. Saffron in phytotherapy: Pharmacology and clinical uses. Wien. Med. Wochenschr. 2007, 157, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Bathaie, S.Z.; Mousavi, S.Z. New applications and mechanisms of action of saffron and its important ingredients. Crit. Rev. Food Sci. Nutr. 2010, 50, 25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, C.-Z.; Wen, X.-D.; Shoyama, Y.; Yuan, C.-S. Role of saffron and its constituents on cancer chemoprevention. Pharm. Biol. 2013, 51, 920–924. [Google Scholar] [CrossRef] [PubMed]
- Castillo, R.; Fernández, J.-A.; Gómez-Gómez, L. Implications of carotenoid biosynthetic genes in apocarotenoid formation during the stigma development of crocus sativus and its closer relatives. Plant Physiol. 2005, 139, 674–689. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Moraga, A.; Ahrazem, O.; Rambla, J.L.; Granell, A.; Gomez-Gomez, L. Crocins with high levels of sugar conjugation contribute to the yellow colours of early-spring flowering crocus tepals. PLoS ONE 2013, 8, e71946. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Rubio-Moraga, A.; Lopez, R.C.; Gomez-Gomez, L. The expression of a chromoplast-specific lycopene β cyclase gene is involved in the high production of saffron’s apocarotenoid precursors. J. Exp. Bot. 2010, 61, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Pfander, H.; Schurtenberger, H. Biosynthesis of c20-carotenoids in Crocus sativus. Phytochemistry 1982, 21, 4. [Google Scholar] [CrossRef]
- Ahrazem, O.; Rubio-Moraga, A.; Berman, J.; Capell, T.; Christou, P.; Zhu, C.; Gómez-Gómez, L. The carotenoid cleavage dioxygenase CCD2 catalyzing the synthesis of crocetin in spring crocuses and saffron is a plastidial enzyme. New Phytol. 2016, 209, 650–663. [Google Scholar] [CrossRef] [PubMed]
- Frusciante, S.; Diretto, G.; Bruno, M.; Ferrante, P.; Pietrella, M.; Prado-Cabrero, A.; Rubio-Moraga, A.; Beyer, P.; Gomez-Gomez, L.; Al-Babili, S.; et al. Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc. Natl. Acad. Sci. USA 2014, 111, 12246–12251. [Google Scholar] [CrossRef] [PubMed]
- Moraga, A.R.; Nohales, P.F.; Perez, J.A.; Gomez-Gomez, L. Glucosylation of the saffron apocarotenoid crocetin by a glucosyltransferase isolated from crocus sativus stigmas. Planta 2004, 219, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Grilli-Caiola, M.; Canini, A. Saffron reproductive biology. Acta Hortic. 2004, 650, 11. [Google Scholar] [CrossRef]
- Bréhélin, C.; Kessler, F.; van Wijk, K.J. Plastoglobules: Versatile lipoprotein particles in plastids. Trends Plant Sci. 2007, 12, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Egea, I.; Barsan, C.; Bian, W.; Purgatto, E.; Latché, A.; Chervin, C.; Bouzayen, M.; Pech, J.-C. Chromoplast differentiation: Current status and perspectives. Plant Cell Physiol. 2010, 51, 1601–1611. [Google Scholar] [CrossRef] [PubMed]
- Rubio, A.; Rambla, J.L.; Santaella, M.; Gómez, M.D.; Orzaez, D.; Granell, A.; Gómez-Gómez, L. Cytosolic and plastoglobule-targeted carotenoid dioxygenases from Crocus sativus are both involved in β-ionone release. J. Biol. Chem. 2008, 283, 24816–24825. [Google Scholar] [CrossRef] [PubMed]
- Nesvizhskii, A.I. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J. Proteom. 2010, 73, 2092–2123. [Google Scholar] [CrossRef] [PubMed]
- Ning, K.; Fermin, D.; Nesvizhskii, A.I. Comparative analysis of different label-free mass spectrometry based protein abundance estimates and their correlation with RNA-seq gene expression data. J. Proteome Res. 2012, 11, 2261–2271. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yin, H.; Lu, H. Recent progress in quantitative glycoproteomics. Glycoconj. J. 2012, 29, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Paredi, G.; Raboni, S.; Marchesani, F.; Ordoudi, S.A.; Tsimidou, M.Z.; Mozzarelli, A.; McPhee, D.J. Insight of saffron proteome by gel-electrophoresis. Molecules 2016, 21, 167. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Sola, M.Á.; Rodríguez-Concepción, M. Carotenoid biosynthesis in arabidopsis: A colorful pathway. Arabidopsis Book 2012, 10, e0158. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-E.; Cheng, K.M.; Craft, N.E.; Hamberger, B.; Douglas, C.J. Over-expression of arabidopsis thaliana carotenoid hydroxylases individually and in combination with a β-carotene ketolase provides insight into in vivo functions. Phytochemistry 2010, 71, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Moraga, A.; Ahrazem, O.; Perez-Clemente, R.M.; Gomez-Cadenas, A.; Yoneyama, K.; Lopez-Raez, J.A.; Molina, R.V.; Gomez-Gomez, L. Apical dominance in saffron and the involvement of the branching enzymes CCD7 and CCD8 in the control of bud sprouting. BMC Plant Biol. 2014, 14, 171. [Google Scholar] [CrossRef] [PubMed]
- Rameau, C. Strigolactones, a novel class of plant hormone controlling shoot branching. C. R. Biol. 2010, 333, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; van Eck, J. Metabolic engineering of carotenoid accumulation by creating a metabolic sink. Transgenic Res. 2007, 16, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Welsch, R.; Yang, Y.; Álvarez, D.; Riediger, M.; Yuan, H.; Fish, T.; Liu, J.; Thannhauser, T.W.; Li, L. Arabidopsis or proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 3558–3563. [Google Scholar] [CrossRef] [PubMed]
- Sitte, P. Development and division of chromoplasts in petals of forsythia. Cellule 1987, 74, 59–77. [Google Scholar]
- Lee, W.D.; Hsu, J.J.; Huang, F.C.; Chao, M.C.; Chang, Y.L.; Huang, M.H. Ischemic stroke in williams-beuren syndrome: A case report. Kaohsiung J. Med. Sci. 2009, 25, 212–216. [Google Scholar] [CrossRef]
- Funes, S.; Gerdes, L.; Inaba, M.; Soll, J.; Herrmann, J.M. The arabidopsis thaliana chloroplast inner envelope protein artemis is a functional member of the Alb3/Oxa1/YidC family of proteins. FEBS Lett. 2004, 569, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Ajjawi, I.; Coku, A.; Froehlich, J.E.; Yang, Y.; Osteryoung, K.W.; Benning, C.; Last, R.L. A j-like protein influences fatty acid composition of chloroplast lipids in arabidopsis. PLoS ONE 2011, 6, e25368. [Google Scholar] [CrossRef] [PubMed]
- Billings, S.E.; Pierzchalski, K.; Tjaden, N.E.B.; Pang, X.-Y.; Trainor, P.A.; Kane, M.A.; Moise, A.R. The retinaldehyde reductase DHRS3 is essential for preventing the formation of excess retinoic acid during embryonic development. FASEB J. 2013, 27, 4877–4889. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Sánchez, V.; Estrada, A.F.; Trautmann, D.; Al-Babili, S.; Avalos, J. The gene carD encodes the aldehyde dehydrogenase responsible for neurosporaxanthin biosynthesis in Fusarium fujikuroi. FEBS J. 2011, 278, 3164–3176. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; McCaffery, P.; Ivins, K.J.; Neve, R.L.; Hogan, P.; Chin, W.W.; Dräger, U.C. Molecular identification of a major retinoic-acid—Synthesizing enzyme, a retinaldehyde—Specific dehydrogenase. Eur. J. Biochem. 1996, 240, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Sánchez, V.; Limón, M.C.; Schaub, P.; Al-Babili, S.; Avalos, J. A raldh-like enzyme involved in fusarium verticillioides development. Fungal Genet. Biol. 2016, 86, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Q.; Yang, Y.; Fei, Z.; Yuan, H.; Fish, T.; Thannhauser, T.W.; Mazourek, M.; Kochian, L.V.; Wang, X.; Li, L. Proteomic analysis of chromoplasts from six crop species reveals insights into chromoplast function and development. J. Exp. Bot. 2013, 64, 949–961. [Google Scholar] [CrossRef] [PubMed]
- Barsan, C.; Sanchez-Bel, P.; Rombaldi, C.; Egea, I.; Rossignol, M.; Kuntz, M.; Zouine, M.; Latche, A.; Bouzayen, M.; Pech, J.C. Characteristics of the tomato chromoplast revealed by proteomic analysis. J. Exp. Bot. 2010, 61, 2413–2431. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, H.; Emes, M. Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Biol. 2000, 51, 111–140. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, F.; Camara, B. The role of plastids in ripening fruits. In The Structure and Function of Plastids; Springer: Dordrecht, The Netherlands, 2006; pp. 419–432. [Google Scholar]
- Cao, H.; Wang, J.; Dong, X.; Han, Y.; Ma, Q.; Ding, Y.; Zhao, F.; Zhang, J.; Chen, H.; Xu, Q. Carotenoid accumulation affects redox status, starch metabolism, and flavonoid/anthocyanin accumulation in citrus. BMC Plant Biol. 2015, 15, 27. [Google Scholar] [CrossRef] [PubMed]
- Kilambi, H.V.; Kumar, R.; Sharma, R.; Sreelakshmi, Y. Chromoplast-specific carotenoid-associated protein appears to be important for enhanced accumulation of carotenoids in hp1 tomato fruits. Plant Physiol. 2013, 161, 2085–2101. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; van Eck, J.; Zhou, X.; Lopez, A.B.; O’Halloran, D.M.; Cosman, K.M.; Conlin, B.J.; Paolillo, D.J.; Garvin, D.F.; Vrebalov, J. The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 2006, 18, 3594–3605. [Google Scholar] [CrossRef] [PubMed]
- Simkin, A.J.; Gaffé, J.; Alcaraz, J.-P.; Carde, J.-P.; Bramley, P.M.; Fraser, P.D.; Kuntz, M. Fibrillin influence on plastid ultrastructure and pigment content in tomato fruit. Phytochemistry 2007, 68, 1545–1556. [Google Scholar] [CrossRef] [PubMed]
- Langenkämper, G.; Manac’h, N.; Broin, M.; Cuiné, S.; Becuwe, N.; Kuntz, M.; Rey, P. Accumulation of plastid lipid-associated proteins (fibrillin/CDSP34) upon oxidative stress, ageing and biotic stress in solanaceae and in response to drought in other species. J. Exp. Bot. 2001, 52, 1545–1554. [Google Scholar] [CrossRef] [PubMed]
- Kreuz, K.; Beyer, P.; Kleinig, H. The site of carotenogenic enzymes in chromoplasts from Narcissus pseudonarcissus L. Planta 1982, 154, 66–69. [Google Scholar] [CrossRef] [PubMed]
- Ljubesić, N.; Wrischer, M.; Devidé, Z. Chromoplasts—The last stages in plastid development. Int. J. Dev. Biol. 1991, 35, 251–258. [Google Scholar] [PubMed]
- Louro, R.P.; Santiago, L.J. Development of carotenoid storage cells in Bixa orellana L. seed arils. Protoplasma 2015, 253, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Weber, R.W.; Davoli, P. Autophagocytosis of carotenoid-rich lipid droplets into vacuoles during aeciospore ageing in Puccinia distincta. New Phytol. 2002, 154, 471–479. [Google Scholar] [CrossRef]
- Corral-Martínez, P.; Parra-Vega, V.; Seguí-Simarro, J.M. Novel features of Brassica napus embryogenic microspores revealed by high pressure freezing and freeze substitution: Evidence for massive autophagy and excretion-based cytoplasmic cleaning. J. Exp. Bot. 2013. [Google Scholar] [CrossRef] [PubMed]
- Seguí-Simarro, J.M. High-pressure freezing and freeze substitution of in vivo and in vitro cultured plant samples. In Plant Microtechniques and Protocols; Yeung, E., Stasolla, C., Sumner, M., Huang, B., Eds.; Springer International Publishing: Switzerland, 2015; pp. 117–134. [Google Scholar]
- Angaman, D.M.; Petrizzo, R.; Hernandez-Gras, F.; Romero-Segura, C.; Pateraki, I.; Busquets, M.; Boronat, A. Precursor uptake assays and metabolic analyses in isolated tomato fruit chromoplasts. Plant Methods 2012, 8, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvachúa, D.; Martínez, A.T.; Tien, M.; López-Lucendo, M.F.; García, F.; de Los Ríos, V.; Martínez, M.J.; Prieto, A. Differential proteomic analysis of the secretome of Irpex lacteus and other white-rot fungi during wheat straw pretreatment. Biotechnol. Biofuels 2013, 6, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diretto, G.; Al-Babili, S.; Tavazza, R.; Scossa, F.; Papacchioli, V.; Migliore, M.; Beyer, P.; Giuliano, G. Transcriptional-metabolic networks in β-carotene-enriched potato tubers: The long and winding road to the golden phenotype. Plant Physiol. 2010, 154, 899–912. [Google Scholar] [CrossRef] [PubMed]








| Uniprot ID | Homologous to Predicted Proteins | Score | Cov. (%) | # Prot. | # UP. | # Pept. | # PSMs. |
|---|---|---|---|---|---|---|---|
| Carotenoid Pathway | |||||||
| H9MZJ1 | 1-Deoxy-d-xylylose 5-phosphate reductoisomerase | 2.9 | 3.9 | 20 | 1 | 1 | 1 |
| J9XGW7 | 4-Hydroxy-3-methylbut-2-enyl diphosphate reductase | 5.56 | 6.74 | 71 | 1 | 2 | 2 |
| W8SQV6 | Aldehyde dehydrogenase 2-1 | 74.4 | 8.6 | 109 | 1 | 4 | 15 |
| A0A061E1Z5 | Aldehyde dehydrogenase 6B2 | 40.4 | 10.0 | 34 | 3 | 4 | 10 |
| M8AU02 | Aldehyde dehydrogenase family 2 member B4 | 28.0 | 5.7 | 34 | 1 | 4 | 8 |
| A0A0B2PSM8 | Aldehyde dehydrogenase family 2 member B7 | 50.3 | 11.0 | 156 | 2 | 6 | 16 |
| Q9LLR2 | Aldehyde dehydrogenase | 111.3 | 10.1 | 99 | 1 | 3 | 25 |
| A0A075W695 | Carotenoid cleavage dioxygenase 2 | 29.0 | 17.3 | 34 | 2 | 8 | 9 |
| A0A075IEG5 | Carotenoid cleavage dioxygenase 7 | 2.6 | 2.7 | 1 | 1 | 1 | 1 |
| Q2R2A6 | Carotenoid isomerase | 48.2 | 5.6 | 27 | 2 | 3 | 11 |
| B7SNW3 | Chromoplast carotenoid cleavage dioxygenase 4b | 11.3 | 7.9 | 5 | 4 | 4 | 4 |
| D2IFC2 | Chromoplast-specific lycopene β cyclase OS=Crocus sativus | 80.4 | 13.4 | 1 | 4 | 4 | 18 |
| Q6X1C0 | Crocetin glucosyltransferase 2 | 24.8 | 10.6 | 18 | 4 | 4 | 6 |
| A0A0D4D933 | CRTISO | 60.7 | 7.7 | 70 | 1 | 4 | 14 |
| S5VPK3 | Lycopene β-cyclase | 8.1 | 10.8 | 65 | 1 | 1 | 2 |
| B9S6H4 | Phytoene dehydrogenase | 16.7 | 2.0 | 1 | 1 | 1 | 4 |
| Q84XU1 | Phytoene desaturase | 105.4 | 25.5 | 229 | 10 | 10 | 26 |
| P37273 | Phytoene synthase 2 | 9.9 | 3.5 | 16 | 1 | 1 | 3 |
| C5I849 | Phytoene synthase | 17.0 | 5.0 | 1 | 1 | 1 | 3 |
| A0A075M6P3 | UDP-glucosyltransferase UGT85U1 | 3.7 | 4.0 | 2 | 1 | 1 | 1 |
| I0AXV3 | ζ-Carotene desaturase | 62.5 | 15.5 | 113 | 2 | 7 | 15 |
| D7TUM8 | ζ-Carotene desaturase | 55.7 | 11.6 | 88 | 1 | 5 | 14 |
| Plastid-Lipid-Associated Proteins | |||||||
| K7UIX3 | Plastid-lipid-associated protein 2 | 15.1 | 9.5 | 19 | 1 | 2 | 4 |
| A0A0B2RM96 | Plastid-lipid-associated protein | 39.0 | 7.8 | 24 | 1 | 2 | 9 |
| A9CSJ8 | Putative plastid lipid-associated protein | 42.7 | 12.2 | 28 | 0 | 2 | 10 |
| Uniprot ID | Homologous to Predicted Proteins | Score | Cov. (%) | # Prot. | # UP. | # Pept. | # PSMs. |
|---|---|---|---|---|---|---|---|
| Carotenoid Pathway | |||||||
| A0A059VDA9 | 1-Deoxy-d-xylulose 5-phosphate reductoisomerase | 39.0 | 24.3 | 95 | 1 | 7 | 10 |
| G8FL07 | 1-Deoxy-d-xylulose 5-phosphate reductoisomerase | 30.0 | 13.6 | 18 | 1 | 5 | 8 |
| A0A075EAM0 | 1-Deoxy-d-xylulose 5-phosphate reductoisomerase | 35.6 | 16.0 | 18 | 1 | 4 | 9 |
| B9RB24 | 1-Deoxy-d-xylulose 5-phosphate reductoisomerase, chloroplast | 15.1 | 4.8 | 3 | 1 | 2 | 4 |
| W6HY22 | Chloroplast 1-deoxy-d-xylulose-5-phosphate reductoisomerase | 49.3 | 24.4 | 133 | 1 | 7 | 13 |
| C7U110 | 1-Deoxy-d-xylulose-5-phosphate synthase 1 | 10.8 | 4.6 | 99 | 1 | 3 | 4 |
| O22567 | 1-Deoxy-d-xylulose-5-phosphate synthase 1, chloroplastic | 11.1 | 4.6 | 94 | 1 | 3 | 4 |
| O81014 | 4-Diphosphocytidyl-2-C-methyl-d-erythritol kinase, chloroplastic | 3.9 | 6.75 | 10 | 1 | 1 | 1 |
| P93841 | 4-Diphosphocytidyl-2-C-methyl-d-erythritol kinase, chloroplastic/chromoplastic | 2.8 | 4.81 | 6 | 1 | 1 | 1 |
| J9XH53 | Putative 4-diphosphocytidyl-2-C-methyl-d-erythritol kinase | 3.8 | 5.24 | 1 | 1 | 1 | 1 |
| D2D5E3 | (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase | 55.3 | 13.78 | 53 | 1 | 7 | 15 |
| W8SNM0 | 4-Hydroxy-3-methylbut-2-en-1-yl diphosphate synthase protein | 54.2 | 10.95 | 72 | 1 | 7 | 15 |
| A0A089G093 | 1-Hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate reductase | 46.3 | 13.61 | 76 | 2 | 6 | 12 |
| W5GTY8 | 2-C-Methyl-d-erythritol 2,4-Cyclodiphosphate synthase | 8.2 | 18.01 | 80 | 3 | 3 | 3 |
| D2CFT5 | Hydroxymethylbutenyl diphosphate reductase | 22.1 | 12.31 | 92 | 2 | 6 | 6 |
| Q9SLG2 | GGPP4_ARATH Geranylgeranyl pyrophosphate synthase 4 | 3.4 | 4.64 | 1 | 1 | 1 | 1 |
| A0A097PPW4 | 15-cis-ζ-carotene isomerase | 7.1 | 4.5 | 1 | 1 | 1 | 2 |
| A0A061E1Z5 | Aldehyde dehydrogenase 6B2 | 40.4 | 11.3 | 36 | 2 | 4 | 8 |
| Q9LRI6 | Aldehyde dehydrogenase ALDH2a | 32.9 | 6.0 | 22 | 1 | 3 | 9 |
| M8AU02 | Aldehyde dehydrogenase family 2 member B4 | 51.2 | 5.7 | 27 | 1 | 4 | 14 |
| W9RMT5 | Aldehyde dehydrogenase family 2 member | 34.1 | 6.4 | 91 | 1 | 3 | 9 |
| A0A059BY89 | Aldehyde dehydrogenase | 13.2 | 7.7 | 85 | 3 | 3 | 4 |
| I1QYB5 | Aldehyde dehydrogenase | 4.2 | 2.3 | 4 | 1 | 1 | 1 |
| Q8VXP2 | β-Carotene hydroxylase | 2.5 | 4.9 | 1 | 1 | 1 | 1 |
| A0A075W695 | Carotenoid cleavage dioxygenase 2 | 72.6 | 23.7 | 1 | 4 | 12 | 18 |
| B7SNW1 | Carotenoid cleavage dioxygenase 2 | 58.6 | 20.8 | 1 | 2 | 10 | 15 |
| V5K7G6 | Carotenoid isomerase (Fragment) | 152.3 | 7.7 | 44 | 1 | 3 | 34 |
| A0A072VJ38 | Carotenoid isomerase | 20.7 | 3.7 | 20 | 1 | 2 | 6 |
| B7SNW3 | Chromoplast carotenoid cleavage dioxygenase 4b | 25.9 | 14.2 | 6 | 6 | 6 | 7 |
| D2IFC2 | Chromoplast-specific lycopene β cyclase | 128.5 | 22.0 | 1 | 7 | 7 | 29 |
| Q6X1C0 | Crocetin glucosyltransferase 2 | 17.8 | 10.6 | 18 | 4 | 4 | 5 |
| M7ZM48 | Cytochrome P450 97B2, chloroplastic | 3.34 | 3.81 | 16 | 1 | 1 | 1 |
| F1BPW8 | Lycopene β-cyclase | 2.9 | 1.8 | 6 | 1 | 1 | 1 |
| S5VPK3 | Lycopene β-cyclase | 19.1 | 10.8 | 65 | 1 | 1 | 5 |
| K9N4H5 | Mitochondrial aldehyde dehydrogenase 2B8 | 68.2 | 7.8 | 155 | 1 | 5 | 20 |
| A0A072TS23 | NAD-dependent aldehyde dehydrogenase family protein | 28.4 | 5.5 | 102 | 1 | 3 | 8 |
| Q84XU1 | Phytoene desaturase | 285.3 | 48.0 | 242 | 17 | 17 | 68 |
| I1U6I7 | Phytoene synthase | 3.6 | 6 | 4 | 1 | 1 | 1 |
| A0A097PKY8 | Phytoene synthase | 12.8 | 6.7 | 195 | 1 | 2 | 4 |
| C5I849 | Phytoene synthase protein | 33.8 | 8.3 | 138 | 1 | 2 | 7 |
| Q52QW5 | Phytoene synthase, chloroplastic | 35.2 | 5.8 | 153 | 1 | 2 | 10 |
| A0A075M6P3 | UDP-glucosyltransferase | 19.7 | 4.0 | 2 | 1 | 1 | 5 |
| I7H187 | ζ-Carotene desaturase | 21.6 | 9.3 | 1 | 1 | 2 | 5 |
| I0AXV3 | ζ-Carotene desaturase | 139.2 | 21.8 | 106 | 5 | 11 | 34 |
| C3VEQ0 | ζ-Carotene desaturase, chloroplastic/chromoplastic | 63.0 | 12.6 | 23 | 2 | 6 | 16 |
| Plastid-Lipid-Associated Proteins | |||||||
| O99019 | Chromoplast-specific carotenoid-associated protein | 12.3 | 10.4 | 1 | 1 | 1 | 3 |
| I1MFL4 | Chromoplast-specific carotenoid-associated protein | 82.8 | 7.8 | 24 | 2 | 2 | 19 |
| R9TH24 | Chromoplast-specific carotenoid-associated protein | 19.2 | 11.2 | 1 | 2 | 2 | 4 |
| Q8S9M1-6 | Isoform 6 of Probable plastid-lipid-associated protein 13, chloroplastic | 2.4 | 5.7 | 21 | 1 | 1 | 1 |
| A6XBI6 | Plastid fibrillin 2 | 7.1 | 4.4 | 7 | 1 | 1 | 2 |
| K7UIX3 | Plastid-lipid-associated protein 2 | 14.7 | 9.5 | 19 | 2 | 2 | 4 |
| A0A075F1U0 | Constitutive plastid-lipid associated protein | 16.5 | 28.9 | 48 | 2 | 3 | 5 |
| K7UIX3 | Plastid-lipid-associated protein 2 | 14.7 | 9.5 | 19 | 2 | 2 | 4 |
| Gene | Pathway | Stigma Developmental Stage | ||||
|---|---|---|---|---|---|---|
| White | Yellow | Orange | Red | Anthesis | ||
| DXS | MEP | 245.6 | 282.5 | 795.7 | 1422.9 | 353.43 |
| DXR | MEP | 58.6 | 56.5 | 119.6 | 282.1 | 26.97 |
| GGPS | MEP | 0 | 11.0 | 51.7 | 13.8 | 0 |
| PSY | Carotenoid | 35.9 | 112.5 | 103.5 | 193.0 | 44.39 |
| PDS | Carotenoid | 52.3 | 76.3 | 228.0 | 189.6 | 46.18 |
| PDS-II | Carotenoid | 4.3 | 8.97 | 10.5 | 0 | 4.44 |
| ZISO | Carotenoid | 110.23 | 135.0 | 319.5 | 186.9 | 217.99 |
| ZDS | Carotenoid | 103 | 139.0 | 436.3 | 568.9 | 1257.168 |
| CTRISO | Carotenoid | 56 | 378.1 | 560.4 | 771.5 | 145.57 |
| LYC-B1 | Carotenoid | 0 | 0 | 16.3 | 17.3 | 0 |
| LYC-B2 | Carotenoid | 63.5 | 111.9 | 465.8 | 407.2 | 55.4 |
| BCH | Carotenoid | 8.7 | 39.4 | 118.2 | 70.6 | 44.28 |
| CCD2 | Apocarotenoid | 93.8 | 307.5 | 693.8 | 594.2 | 15.83 |
| UGT2 | Apocarotenoid | 68.2 | 943.6 | 1567.2 | 1935.4 | 256.6 |
| UGT85U1 | Apocarotenoid | 62.1 | 218.9 | 688.9 | 541.6 | 1579.1 |
| CCD7 | Apocarotenoid | 0 | 0 | 23.5 | 0 | 0 |
| CCD4b | Apocarotenoid | 0 | 0 | 0 | 11.7 | 46.5 |
| ADHComp2946 | Apocarotenoid | 17.6 | 254.6 | 321.4 | 575.2 | 561.6 |
| ADHComp20158 | Apocarotenoid | 171.2 | 60.4 | 54.5 | 69.7 | 0 |
| ADHComp3898 | Apocarotenoid | 34.5 | 90.3 | 90.9 | 29.1 | 29.6 |
| ADHComp54788 | Apocarotenoid | 35.5 | 57.4 | 134.8 | 132.5 | 319.6 |
| ADHComp11367 | Apocarotenoid | 37.3 | 25.4 | 61.6 | 101.4 | 66.2 |
| GLUCOSEPYROPHOSPHORYLASE (UGP3) | Starch | 80.1 | 207.4 | 196.9 | 166.7 | 29.3 |
| STARCH BRANCHING ENZYME (SBE2.1) | Starch | 8.4 | 16.3 | 23 | 40.7 | 3.2 |
| STARCH BRANCHING ENZYME (SBE2.2) | Starch | 14.2 | 26.5 | 38.8 | 24.8 | 0 |
| GRANULE BOUND STARCH SYNTHASE (GBSS1) | Starch | 56.4 | 116.2 | 108 | 25.9 | 4 |
| STARCH SYNTHASE (SS2) | Starch | 60.3 | 41.1 | 0 | 9.5 | 0 |
| STARCH SYNTHASE (SS3) | Starch | 58.9 | 58.5 | 32 | 31.4 | 8 |
| B-AMILASE3 | Starch | 4.8 | 5 | 23.5 | 14.4 | 74.6 |
| ALFA AMILASE (ISA3) | Starch | 19.7 | 16 | 9.4 | 9.9 | 10 |
| ALFA AMILASE (AMY3) | Starch | 2.9 | 6 | 7.1 | 0 | 0 |
| ALFA AMILASE (LDA) | Starch | 4.2 | 9.9 | 0 | 12.3 | 14.6 |
| Plastid fibrillin 2 | Plastid development | 20.5 | 62.1 | 146.3 | 95 | 75.3 |
| Plastid-lipid-associated protein 2 | Plastid development | 4.1 | 17 | 0 | 117 | 50.6 |
| Isoform 6 of Probable plastid-lipid-associated protein 13 | Plastid development | 13.6 | 41.2 | 161.4 | 94.6 | 6.8 |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Gómez, L.; Parra-Vega, V.; Rivas-Sendra, A.; Seguí-Simarro, J.M.; Molina, R.V.; Pallotti, C.; Rubio-Moraga, Á.; Diretto, G.; Prieto, A.; Ahrazem, O. Unraveling Massive Crocins Transport and Accumulation through Proteome and Microscopy Tools during the Development of Saffron Stigma. Int. J. Mol. Sci. 2017, 18, 76. https://doi.org/10.3390/ijms18010076
Gómez-Gómez L, Parra-Vega V, Rivas-Sendra A, Seguí-Simarro JM, Molina RV, Pallotti C, Rubio-Moraga Á, Diretto G, Prieto A, Ahrazem O. Unraveling Massive Crocins Transport and Accumulation through Proteome and Microscopy Tools during the Development of Saffron Stigma. International Journal of Molecular Sciences. 2017; 18(1):76. https://doi.org/10.3390/ijms18010076
Chicago/Turabian StyleGómez-Gómez, Lourdes, Verónica Parra-Vega, Alba Rivas-Sendra, Jose M. Seguí-Simarro, Rosa Victoria Molina, Claudia Pallotti, Ángela Rubio-Moraga, Gianfranco Diretto, Alicia Prieto, and Oussama Ahrazem. 2017. "Unraveling Massive Crocins Transport and Accumulation through Proteome and Microscopy Tools during the Development of Saffron Stigma" International Journal of Molecular Sciences 18, no. 1: 76. https://doi.org/10.3390/ijms18010076
APA StyleGómez-Gómez, L., Parra-Vega, V., Rivas-Sendra, A., Seguí-Simarro, J. M., Molina, R. V., Pallotti, C., Rubio-Moraga, Á., Diretto, G., Prieto, A., & Ahrazem, O. (2017). Unraveling Massive Crocins Transport and Accumulation through Proteome and Microscopy Tools during the Development of Saffron Stigma. International Journal of Molecular Sciences, 18(1), 76. https://doi.org/10.3390/ijms18010076