Evolutionary Conserved Cysteines Function as cis-Acting Regulators of Arabidopsis PIN-FORMED 2 Distribution
Abstract
:1. Introduction
2. Results
2.1. PIN Proteins Share Conserved Cysteines that Contribute to Protein Functionality
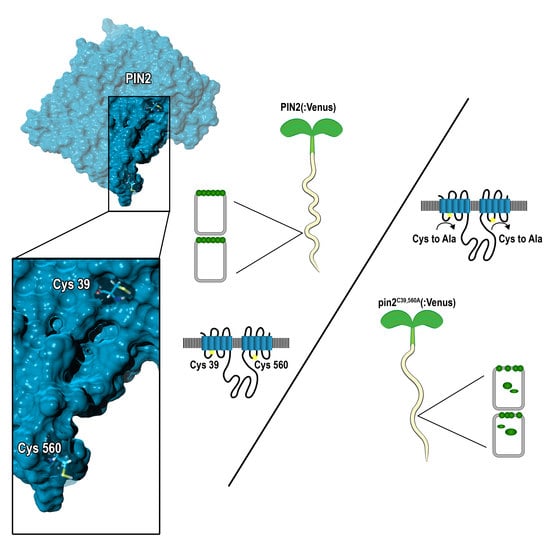
2.2. Conserved Cysteines Are Determinants of PIN2 Intracellular Distribution
3. Discussion
4. Materials and Methods
4.1. Plant Lines, Growth Conditions and Vector Construction
4.2. Microscopy
4.3. Homology Modeling and Molecular Dynamics
4.4. Data Acquisition and Processing
4.5. PIN Sequences and Alignments
4.6. Expression Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lavy, M.; Estelle, M. Mechanisms of auxin signaling. Development 2016, 143, 3226–3229. [Google Scholar] [CrossRef] [PubMed]
- Enders, T.A.; Strader, L.C. Auxin activity: Past, present, and future. Am. J. Bot. 2015, 102, 180–196. [Google Scholar] [CrossRef] [PubMed]
- Leyser, O. Auxin signaling. Plant Physiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Adamowski, M.; Friml, J. PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Borghi, L.; Kang, J.; Ko, D.; Lee, Y.; Martinoia, E. The role of ABCG-type ABC transporters in phytohormone transport. Biochem. Soc. Trans. 2015, 43, 924–930. [Google Scholar] [CrossRef] [PubMed]
- Zazimalova, E.; Murphy, A.S.; Yang, H.; Hoyerova, K.; Hosek, P. Auxin transporters—Why so many? Cold Spring Harb Perspect Biol 2010, 2, a001552. [Google Scholar] [CrossRef] [PubMed]
- Luschnig, C.; Vert, G. The dynamics of plant plasma membrane proteins: PINs and beyond. Development 2014, 141, 2924–2938. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Zago, M.K.; Abas, L.; van Marion, A.; Galvan-Ampudia, C.S.; Offringa, R. Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 2010, 22, 1129–1142. [Google Scholar] [CrossRef] [PubMed]
- Zourelidou, M.; Absmanner, B.; Weller, B.; Barbosa, I.C.; Willige, B.C.; Fastner, A.; Streit, V.; Port, S.A.; Colcombet, J.; de la Fuente van Bentem, S.; et al. Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. Elife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Michniewicz, M.; Zago, M.K.; Abas, L.; Weijers, D.; Schweighofer, A.; Meskiene, I.; Heisler, M.G.; Ohno, C.; Zhang, J.; Huang, F.; et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 2007, 130, 1044–1056. [Google Scholar] [CrossRef] [PubMed]
- Korbei, B.; Luschnig, C. Plasma membrane protein ubiquitylation and degradation as determinants of positional growth in plants. J. Integr. Plant Biol. 2013, 55, 809–823. [Google Scholar] [CrossRef] [PubMed]
- Isono, E.; Kalinowska, K. ESCRT-dependent degradation of ubiquitylated plasma membrane proteins in plants. Curr. Opin. Plant Biol. 2017, 40, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Korbei, B.; Moulinier-Anzola, J.; De-Araujo, L.; Lucyshyn, D.; Retzer, K.; Khan, M.A.; Luschnig, C. Arabidopsis TOL proteins act as gatekeepers for vacuolar sorting of PIN2 plasma membrane protein. Curr. Biol. 2013, 23, 2500–2505. [Google Scholar] [CrossRef] [PubMed]
- Nagel, M.K.; Kalinowska, K.; Vogel, K.; Reynolds, G.D.; Wu, Z.X.; Anzenberger, F.; Ichikawa, M.; Tsutsumi, C.; Sato, M.H.; Kuster, B.; et al. Arabidopsis SH3P2 is an ubiquitin-binding protein that functions together with ESCRT-I and the deubiquitylating enzyme AMSH3. Proc. Natl. Acad. Sci. USA 2017, 114, E7197–E7204. [Google Scholar] [CrossRef] [PubMed]
- Leitner, J.; Petrasek, J.; Tomanov, K.; Retzer, K.; Parezova, M.; Korbei, B.; Bachmair, A.; Zazimalova, E.; Luschnig, C. Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc. Natl. Acad. Sci. USA 2012, 109, 8322–8327. [Google Scholar] [CrossRef] [PubMed]
- Giles, N.M.; Watts, A.B.; Giles, G.I.; Fry, F.H.; Littlechild, J.A.; Jacob, C. Metal and redox modulation of cysteine protein function. Chem. Biol. 2003, 10, 677–693. [Google Scholar] [CrossRef]
- Xia, X.J.; Zhou, Y.H.; Shi, K.; Zhou, J.; Foyer, C.H.; Yu, J.Q. Interplay between reactive oxygen species and hormones in the control of plant. J. Exp. Bot. 2015, 66, 2839–2856. [Google Scholar] [CrossRef] [PubMed]
- Rouhier, N.; Cerveau, D.; Couturier, J.; Reichheld, J.P.; Rey, P. Involvement of thiol-based mechanisms in plant development. Biochim. Biophys. Acta 2015, 1850, 1479–1496. [Google Scholar] [CrossRef] [PubMed]
- Bashandy, T.; Guilleminot, J.; Vernoux, T.; Caparros-Ruiz, D.; Ljung, K.; Meyer, Y.; Reichheld, J.P. Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 2010, 22, 376–391. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Pasternak, T.; Eiblmeier, M.; Ditengou, F.; Kochersperger, P.; Sun, J.; Wang, H.; Rennenberg, H.; Teale, W.; Paponov, I.; et al. Plastid-localized glutathione reductase2-regulated glutathione redox status is essential for Arabidopsis root apical meristem maintenance. Plant Cell 2013, 25, 4451–4468. [Google Scholar] [CrossRef] [PubMed]
- Koprivova, A.; Mugford, S.T.; Kopriva, S. Arabidopsis root growth dependence on glutathione is linked to auxin transport. Plant Cell Rep. 2010, 29, 1157–1167. [Google Scholar] [CrossRef] [PubMed]
- Feechan, A.; Kwon, E.; Yun, B.W.; Wang, Y.; Pallas, J.A.; Loake, G.J. A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. USA 2005, 102, 8054–8059. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.F.; Wang, D.L.; Wang, C.; Culler, A.H.; Kreiser, M.A.; Suresh, J.; Cohen, J.D.; Pan, J.; Baker, B.; Liu, J.Z. Loss of GSNOR1 function leads to compromised auxin signaling and polar auxin transport. Mol. Plant 2015, 8, 1350–1365. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Marcos, M.; Sanz, L.; Lewis, D.R.; Muday, G.K.; Lorenzo, O. Nitric oxide causes root apical meristem defects and growth inhibition while reducing PIN-FORMED 1 (PIN1)-dependent acropetal auxin transport. Proc. Natl. Acad. Sci. USA 2011, 108, 18506–18511. [Google Scholar] [CrossRef] [PubMed]
- Luschnig, C.; Gaxiola, R.A.; Grisafi, P.; Fink, G.R. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 1998, 12, 2175–2187. [Google Scholar] [CrossRef] [PubMed]
- Krecek, P.; Skupa, P.; Libus, J.; Naramoto, S.; Tejos, R.; Friml, J.; Zazimalova, E. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009, 10, 249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, T.; Brockington, S.F.; Rothfels, C.; Graham, S.W.; Stevenson, D.; Kutchan, T.; Rolf, M.; Thomas, P.; Wong, G.K.; Leyser, O.; et al. Paralogous radiations of PIN proteins with multiple origins of noncanonical PIN structure. Mol. Biol. Evol. 2014, 31, 2042–2060. [Google Scholar] [CrossRef] [PubMed]
- Nodzynski, T.; Vanneste, S.; Zwiewka, M.; Pernisova, M.; Hejatko, J.; Friml, J. Enquiry into the topology of plasma membrane-localized PIN auxin transport components. Mol. Plant 2016, 9, 1504–1519. [Google Scholar] [CrossRef] [PubMed]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Cerutti, D.S.; Cheatham, I.T.E.; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Greene, D.; Homeyer, N.; et al. AMBER 2017; University of California: San Francisco, CA, USA, 2017. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Grabov, A.; Ashley, M.K.; Rigas, S.; Hatzopoulos, P.; Dolan, L.; Vicente-Agullo, F. Morphometric analysis of root shape. New Phytol. 2005, 165, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Kleine-Vehn, J.; Wabnik, K.; Martiniere, A.; Langowski, L.; Willig, K.; Naramoto, S.; Leitner, J.; Tanaka, H.; Jakobs, S.; Robert, S.; et al. Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol. Syst. Biol. 2011, 7, 540. [Google Scholar] [CrossRef] [PubMed]
- Martiniere, A.; Lavagi, I.; Nageswaran, G.; Rolfe, D.J.; Maneta-Peyret, L.; Luu, D.T.; Botchway, S.W.; Webb, S.E.; Mongrand, S.; Maurel, C.; et al. Cell wall constrains lateral diffusion of plant plasma-membrane proteins. Proc. Natl. Acad. Sci. USA 2012, 109, 12805–12810. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, N.; Culley, S.; Ashdown, G.; Owen, D.M.; Pereira, P.M.; Henriques, R. Fast live-cell conventional fluorophore nanoscopy with ImageJ through super-resolution radial fluctuations. Nat. Commun. 2016, 7, 12471. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, A.S.; Chaumont, F. Trafficking of plant plasma membrane aquaporins: Multiple regulation levels and complex sorting signals. Plant Cell Physiol. 2015, 56, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Bienert, G.P.; Cavez, D.; Besserer, A.; Berny, M.C.; Gilis, D.; Rooman, M.; Chaumont, F. A conserved cysteine residue is involved in disulfide bond formation between plant plasma membrane aquaporin monomers. Biochem. J. 2012, 445, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.J.; Lee, H.K.; Han, W.; Kim, D.H.; Lee, M.H.; Jeon, J.; Lee, D.W.; Lee, J.; Lee, Y.; Lee, J.; et al. Interactions between transmembrane helices within monomers of the aquaporin AtPIP2;1 play a crucial role in tetramer formation. Mol. Plant 2016, 9, 1004–1017. [Google Scholar] [CrossRef] [PubMed]
- Mou, Z.; Fan, W.; Dong, X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Despres, C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Feraru, E.; Feraru, M.I.; Kleine-Vehn, J.; Martiniere, A.; Mouille, G.; Vanneste, S.; Vernhettes, S.; Runions, J.; Friml, J. PIN polarity maintenance by the cell wall in Arabidopsis. Curr. Biol. 2011, 21, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Van Breusegem, F. ROS signaling: The new wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Abas, L.; Benjamins, R.; Malenica, N.; Paciorek, T.; Wisniewska, J.; Moulinier-Anzola, J.C.; Sieberer, T.; Friml, J.; Luschnig, C. Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 2006, 8, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Koncz, C.; Schell, J. The promoter of Tl-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 1986, 204, 383–396. [Google Scholar] [CrossRef]
- Jones, D.T. GenTHREADER: An efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol. 1999, 287, 797–815. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [PubMed]
- McGuffin, L.J.; Jones, D.T. Improvement of the GenTHREADER method for genomic fold recognition. Bioinformatics 2003, 19, 874–881. [Google Scholar] [CrossRef] [PubMed]
- Lobley, A.; Sadowski, M.I.; Jones, D.T. pGenTHREADER and pDomTHREADER: New methods for improved protein fold recognition and superfamily discrimination. Bioinformatics 2009, 25, 1761–1767. [Google Scholar] [CrossRef] [PubMed]
- Mongan, J.; Simmerling, C.; McCammon, J.A.; Case, D.A.; Onufriev, A. Generalized Born model with a simple, robust molecular volume correction. J. Chem. Theory Comput. 2007, 3, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.; Roe, D.R.; Simmerling, C. Improved generalized born solvent model parameters for protein simulations. J. Chem. Theory Comput. 2013, 9, 2020–2034. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins 2006, 65, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical-integration of Cartesian equations of motion of a system with constraints—Molecular-dynamics of N-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. Settle—An analytical version of the Shake and Rattle algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Leinonen, R.; Sugawara, H.; Shumway, M.; International Nucleotide Sequence Database Collaboration. The sequence read archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Hellemans, J.; Mortier, G.; de Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed]








© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Retzer, K.; Lacek, J.; Skokan, R.; Del Genio, C.I.; Vosolsobě, S.; Laňková, M.; Malínská, K.; Konstantinova, N.; Zažímalová, E.; Napier, R.M.; et al. Evolutionary Conserved Cysteines Function as cis-Acting Regulators of Arabidopsis PIN-FORMED 2 Distribution. Int. J. Mol. Sci. 2017, 18, 2274. https://doi.org/10.3390/ijms18112274
Retzer K, Lacek J, Skokan R, Del Genio CI, Vosolsobě S, Laňková M, Malínská K, Konstantinova N, Zažímalová E, Napier RM, et al. Evolutionary Conserved Cysteines Function as cis-Acting Regulators of Arabidopsis PIN-FORMED 2 Distribution. International Journal of Molecular Sciences. 2017; 18(11):2274. https://doi.org/10.3390/ijms18112274
Chicago/Turabian StyleRetzer, Katarzyna, Jozef Lacek, Roman Skokan, Charo I. Del Genio, Stanislav Vosolsobě, Martina Laňková, Kateřina Malínská, Nataliia Konstantinova, Eva Zažímalová, Richard M. Napier, and et al. 2017. "Evolutionary Conserved Cysteines Function as cis-Acting Regulators of Arabidopsis PIN-FORMED 2 Distribution" International Journal of Molecular Sciences 18, no. 11: 2274. https://doi.org/10.3390/ijms18112274
APA StyleRetzer, K., Lacek, J., Skokan, R., Del Genio, C. I., Vosolsobě, S., Laňková, M., Malínská, K., Konstantinova, N., Zažímalová, E., Napier, R. M., Petrášek, J., & Luschnig, C. (2017). Evolutionary Conserved Cysteines Function as cis-Acting Regulators of Arabidopsis PIN-FORMED 2 Distribution. International Journal of Molecular Sciences, 18(11), 2274. https://doi.org/10.3390/ijms18112274