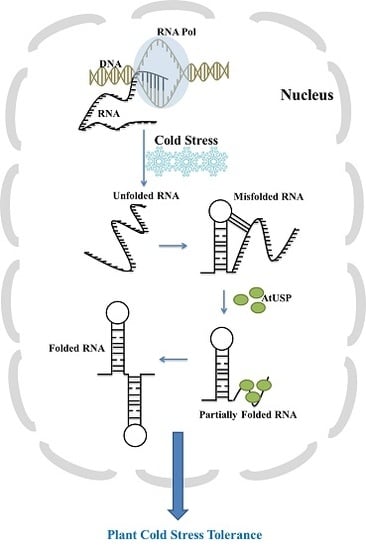
RNA Chaperone Function of a Universal Stress Protein in Arabidopsis Confers Enhanced Cold Stress Tolerance in Plants
Abstract
:1. Introduction
2. Results
2.1. Transgenic Arabidopsis Overexpressing AtUSP Displays Enhanced Resistance to Chilling and Freezing Stress
2.2. Subcellular Localization of AtUSP
2.3. DNA- and RNA-Binding Activity of AtUSP
2.4. Nucleic Acid-Melting Activity of AtUSP
2.5. AtUSP Exhibits Anti-Termination Activity in the Transcription of Bacterial Chloramphenicol Acetyltransferase (CAT)
2.6. AtUSP Complements Cold-Shock Sensitive BX04 E. coli Mutant Cells
3. Discussion
4. Materials and Methods
4.1. RT-PCR and qPCR Analysis of mRNA Expression
4.2. Chilling and Freezing Tolerance Assay
4.3. Electrolyte Leakage Test
4.4. Subcellular Localization of AtUSP
4.5. Purification of AtUSP Recombinant Protein
4.6. Nucleic Acid-Binding Analysis
4.7. DNA-Melting Assay
4.8. Transcription Anti-Termination and Cold Complementation Assay in E. coli
4.9. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Baurle, I. Plant Heat Adaptation: Priming in response to heat stress. F1000Research 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Burce, T.J.A.; Matthes, M.C.; Napier, J.A. Stressful “memories” of plants: Evidence and possible mechanisms. Plant Sci. Int. J. Exp. Plant Biol. 2007, 173, 603–608. [Google Scholar] [CrossRef]
- Conrath, U. Molecular aspects of defence priming. Trends Plant Sci. 2011, 16, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Kvint, K.; Nachin, L.; Diez, A.; Nystrom, T. The bacterial universal stress protein: Function and regulation. Curr. Opin. Microbiol. 2003, 6, 140–145. [Google Scholar] [CrossRef]
- Nystrom, T.; Neidhardt, F.C. Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol. Microbiol. 1992, 6, 3187–3198. [Google Scholar] [CrossRef] [PubMed]
- Siegele, D.A. Universal stress proteins in Escherichia coli. J. Bacteriol. 2005, 187, 6253–6254. [Google Scholar] [CrossRef] [PubMed]
- Nachin, L.; Nannmark, U.; Nystrom, T. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 2005, 187, 6265–6272. [Google Scholar] [CrossRef] [PubMed]
- Sousa, M.C.; McKay, D.B. Structure of the universal stress protein of Haemophilus influenzae. Structure 2001, 9, 1135–1141. [Google Scholar] [CrossRef]
- Zarembinski, T.I.; Hung, L.W.; Mueller-Dieckmann, H.J.; Kim, K.K.; Yokota, H.; Kim, R.; Kim, S.H. Structure-based assignment of the biochemical function of a hypothetical protein: A test case of structural genomics. Proc. Natl. Acad. Sci. USA 1998, 95, 15189–15193. [Google Scholar] [CrossRef] [PubMed]
- Isokpehi, R.D.; Mahmud, O.; Mbah, A.N.; Simmons, S.S.; Avelar, L.; Rajnarayanan, R.V.; Udensi, U.K.; Ayensu, W.K.; Cohly, H.H.; Brown, S.D.; et al. Developmental Regulation of Genes Encoding Universal Stress Proteins in Schistosoma mansoni. Gene Regul. Syst. Biol. 2011, 5, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Kerk, D.; Bulgrien, J.; Smith, D.W.; Gribskov, M. Arabidopsis proteins containing similarity to the universal stress protein domain of bacteria. Plant Physiol. 2003, 131, 1209–1219. [Google Scholar] [CrossRef] [PubMed]
- Isokpehi, R.D.; Simmons, S.S.; Cohly, H.H.; Ekunwe, S.I.; Begonia, G.B.; Ayensu, W.K. Identification of drought-responsive universal stress proteins in viridiplantae. Bioinform. Biol. Insights 2011, 5, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Sauter, M.; Rzewuski, G.; Marwedel, T.; Lorbiecke, R. The novel ethylene-regulated gene OsUsp1 from rice encodes a member of a plant protein family related to prokaryotic universal stress proteins. J. Exp. Bot. 2002, 53, 2325–2331. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Chou, M.X.; Wei, X.Y.; Chen, D.S.; Zhou, J.C. A novel nodule-enhanced gene encoding a putative universal stress protein from Astragalus sinicus. J. Plant Physiol. 2007, 164, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Gonzali, S.; Loreti, E.; Cardarelli, F.; Novi, G.; Parlanti, S.; Pucciariello, C.; Bassolino, L.; Banti, V.; Licausi, F.; Perata, P. Universal stress protein HRU1 mediates ROS homeostasis under anoxia. Nat. Plants 2015, 1, 15151. [Google Scholar] [CrossRef] [PubMed]
- Loukehaich, R.; Wang, T.; Ouyang, B.; Ziaf, K.; Li, H.; Zhang, J.; Lu, Y.; Ye, Z. SpUSP, an annexin-interacting universal stress protein, enhances drought tolerance in tomato. J. Exp. Bot. 2012, 63, 5593–5606. [Google Scholar] [CrossRef] [PubMed]
- Udawat, P.; Mishra, A.; Jha, B. Heterologous expression of an uncharacterized universal stress protein gene (SbUSP) from the extreme halophyte, Salicornia brachiata, which confers salt and osmotic tolerance to E. coli. Gene 2014, 536, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.J.; Melencion, S.M.; Lee, E.S.; Park, J.H.; Alinapon, C.V.; Oh, H.T.; Yun, D.J.; Chi, Y.H.; Lee, S.Y. Universal Stress Protein Exhibits a Redox-Dependent Chaperone Function in Arabidopsis and Enhances Plant Tolerance to Heat Shock and Oxidative Stress. Front. Plant Sci. 2015, 6, 1141. [Google Scholar] [CrossRef] [PubMed]
- Chae, H.B.; Moon, J.C.; Shin, M.R.; Chi, Y.H.; Jung, Y.J.; Lee, S.Y.; Nawkar, G.M.; Jung, H.S.; Hyun, J.K.; Kim, W.Y.; et al. Thioredoxin reductase type C (NTRC) orchestrates enhanced thermotolerance to Arabidopsis by its redox-dependent holdase chaperone function. Mol. Plant 2013, 6, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.H.; Paeng, S.K.; Kim, M.J.; Hwang, G.Y.; Melencion, S.M.; Oh, H.T.; Lee, S.Y. Redox-dependent functional switching of plant proteins accompanying with their structural changes. Front. Plant Sci. 2013, 4, 277. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.H.; Lee, K.O.; Chi, Y.H.; Jung, B.G.; Park, S.K.; Park, J.H.; Lee, J.R.; Lee, S.S.; Moon, J.C.; Yun, J.W.; et al. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 2004, 117, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, Y.; Uemura, M. Mass spectrometric approach for identifying putative plasma membrane proteins of Arabidopsis leaves associated with cold acclimation. Plant J. Cell Mol. Biol. 2003, 36, 141–154. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Dong, C.H.; Lee, H.; Zhu, J.; Xiong, L.; Gong, D.; Stevenson, B.; Zhu, J.K. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 2005, 17, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Drumm, J.E.; Mi, K.; Bilder, P.; Sun, M.; Lim, J.; Bielefeldt-Ohmann, H.; Basaraba, R.; So, M.; Zhu, G.; Tufariello, J.M.; et al. Mycobacterium tuberculosis universal stress protein Rv2623 regulates bacillary growth by ATP-Binding: Requirement for establishing chronic persistent infection. PLoS Pathog. 2009, 5, e1000460. [Google Scholar] [CrossRef]
- Sasaki, K.; Kim, M.H.; Imai, R. Arabidopsis COLD SHOCK DOMAIN PROTEIN2 is a RNA chaperone that is regulated by cold and developmental signals. Biochem. Biophys. Res. Commun. 2007, 364, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Diez, A.; Gustavsson, N.; Nystrom, T. The universal stress protein A of Escherichia coli is required for resistance to DNA damaging agents and is regulated by a RecA/FtsK-dependent regulatory pathway. Mol. Microbiol. 2000, 36, 1494–1503. [Google Scholar] [CrossRef] [PubMed]
- Gendra, E.; Moreno, A.; Alba, M.M.; Pages, M. Interaction of the plant glycine-rich RNA-binding protein MA16 with a novel nucleolar DEAD box RNA helicase protein from Zea mays. Plant J. Cell Mol. Biol. 2004, 38, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kim, W.Y.; Kwak, K.J.; Oh, S.H.; Han, Y.S.; Kang, H. Glycine-rich RNA-binding proteins are functionally conserved in Arabidopsis thaliana and Oryza sativa during cold adaptation process. J. Exp. Bot. 2010, 61, 2317–2325. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kim, W.Y.; Kwak, K.J.; Oh, S.H.; Han, Y.S.; Kang, H. Zinc finger-containing glycine-rich RNA-binding protein in Oryza sativa has an RNA chaperone activity under cold stress conditions. Plant Cell Environ. 2010, 33, 759–768. [Google Scholar] [PubMed]
- Kim, J.Y.; Park, S.J.; Jang, B.; Jung, C.H.; Ahn, S.J.; Goh, C.H.; Cho, K.; Han, O.; Kang, H. Functional characterization of a glycine-rich RNA-binding protein 2 in Arabidopsis thaliana under abiotic stress conditions. Plant J. Cell Mol. Biol. 2007, 50, 4351–4394. [Google Scholar] [CrossRef] [PubMed]
- Landick, R.; Stewart, J.; Lee, D.N. Amino acid changes in conserved regions of the beta-subunit of Escherichia coli RNA polymerase alter transcription pausing and termination. Genes Dev. 1990, 4, 1623–1636. [Google Scholar] [CrossRef] [PubMed]
- Phadtare, S.; Severinov, K. Nucleic acid melting by Escherichia coli CspE. Nucleic Acids Res. 2005, 33, 5583–5590. [Google Scholar] [CrossRef] [PubMed]
- Rajkowitsch, L.; Semrad, K.; Mayer, O.; Schroeder, R. Assays for the RNA chaperone activity of proteins. Biochem. Soc. Trans. 2005, 33, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Hou, Y.; Inouye, M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 1997, 272, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Cristofari, G.; Darlix, J.L. The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acid Res. Mol. Biol. 2002, 72, 223–268. [Google Scholar] [PubMed]
- Herschlag, D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995, 270, 20871–20874. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, R.; Barta, A.; Semrad, K. Strategies for RNA folding and assembly. Nat. Rev. Mol. Cell Biol. 2004, 5, 908–919. [Google Scholar] [CrossRef] [PubMed]
- Semrad, K. Proteins with RNA chaperone activity: A world of diverse proteins with a common task-impediment of RNA misfolding. Biochem. Res. Int. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Phadtare, S.; Inouye, M.; Severinov, K. The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J. Biol. Chem. 2002, 277, 7239–7245. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, J.M.; Lee, H.N.; Kim, E.K.; Ha, B.; Ahn, S.M.; Jang, H.H.; Lee, S.Y. RNA-binding properties and RNA chaperone activity of human peroxiredoxin 1. Biochem. Biophys. Res. Commun. 2012, 425, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.O.; Kang, H. The role of a zinc finger-containing glycine-rich RNA-binding protein during the cold adaptation process in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.O.; Kim, J.S.; Kang, H. Cold-inducible zinc finger-containing glycine-rich RNA-binding protein contributes to the enhancement of freezing tolerance in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2005, 42, 890–900. [Google Scholar] [CrossRef] [PubMed]
- Bhuria, M.; Goel, P.; Kumar, S.; Singh, A.K. The Promoter of AtUSP Is Co-regulated by Phytohormones and Abiotic Stresses in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 1957. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Shi, Y.; Liu, J.; Guo, L.; Zhang, X.; Yang, S. A mutant CHS3 protein with TIR-NB-LRR-LIM domains modulates growth, cell death and freezing tolerance in a temperature-dependent manner in Arabidopsis. Plant J. Cell Mol. Biol. 2010, 63, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Kim, D.H.; Kim, Y.-W.; Hwang, I. Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 2001, 13, 2175–2190. [Google Scholar] [CrossRef] [PubMed]








© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melencion, S.M.B.; Chi, Y.H.; Pham, T.T.; Paeng, S.K.; Wi, S.D.; Lee, C.; Ryu, S.W.; Koo, S.S.; Lee, S.Y. RNA Chaperone Function of a Universal Stress Protein in Arabidopsis Confers Enhanced Cold Stress Tolerance in Plants. Int. J. Mol. Sci. 2017, 18, 2546. https://doi.org/10.3390/ijms18122546
Melencion SMB, Chi YH, Pham TT, Paeng SK, Wi SD, Lee C, Ryu SW, Koo SS, Lee SY. RNA Chaperone Function of a Universal Stress Protein in Arabidopsis Confers Enhanced Cold Stress Tolerance in Plants. International Journal of Molecular Sciences. 2017; 18(12):2546. https://doi.org/10.3390/ijms18122546
Chicago/Turabian StyleMelencion, Sarah Mae Boyles, Yong Hun Chi, Thuy Thi Pham, Seol Ki Paeng, Seong Dong Wi, Changyu Lee, Seoung Woo Ryu, Sung Sun Koo, and Sang Yeol Lee. 2017. "RNA Chaperone Function of a Universal Stress Protein in Arabidopsis Confers Enhanced Cold Stress Tolerance in Plants" International Journal of Molecular Sciences 18, no. 12: 2546. https://doi.org/10.3390/ijms18122546
APA StyleMelencion, S. M. B., Chi, Y. H., Pham, T. T., Paeng, S. K., Wi, S. D., Lee, C., Ryu, S. W., Koo, S. S., & Lee, S. Y. (2017). RNA Chaperone Function of a Universal Stress Protein in Arabidopsis Confers Enhanced Cold Stress Tolerance in Plants. International Journal of Molecular Sciences, 18(12), 2546. https://doi.org/10.3390/ijms18122546