LncRNA Structural Characteristics in Epigenetic Regulation
Abstract
:1. Introduction
2. The Genome Architecture of LncRNAs
3. The Epigenetic Regulatory Activity of lncRNAs
4. The Structural Basement of LncRNA in Epigenetic Regulation
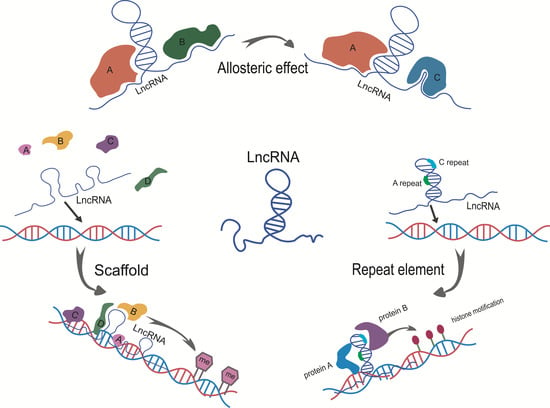
5. The Models of LncRNA Structure Mediated Epigenetic Regulation
6. Summary and Expectation
Acknowledgments
Conflicts of Interest
References
- Schmitt, A.M.; Garcia, J.T.; Hung, T.; Flynn, R.A.; Shen, Y.; Qu, K.; Payumo, A.Y.; Peres-da-Silva, A.; Broz, D.K.; Baum, R.; et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat. Genet. 2016, 48, 1370–1376. [Google Scholar] [CrossRef] [PubMed]
- Rabani, M.; Raychowdhury, R.; Jovanovic, M.; Rooney, M.; Stumpo, D.J.; Pauli, A.; Hacohen, N.; Schier, A.F.; Blackshear, P.J.; Friedman, N.; et al. High-resolution sequencing and modeling identifies distinct dynamic RNA regulatory strategies. Cell 2014, 159, 1698–1710. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, Q.; Wang, X.; Zheng, J.; Wang, T.; You, M.; Sheng Sun, Z.; Shi, Q. mirTools 2.0 for non-coding RNA discovery, profiling, and functional annotation based on high-throughput sequencing. RNA Biol. 2013, 10, 1087–1092. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T. Epigenetic regulation by long noncoding RNAs. Science 2012, 338, 1435–1439. [Google Scholar] [CrossRef] [PubMed]
- Bu, D.; Yu, K.; Sun, S.; Xie, C.; Skogerbo, G.; Miao, R.; Xiao, H.; Liao, Q.; Luo, H.; Zhao, G.; et al. NONCODE v3.0: Integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2012, 40, D210–D215. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Su, Y.; Wu, H.; Dai, Y.; Zhao, M.; Lu, Q. Characters, functions and clinical perspectives of long non-coding RNAs. Mol. Genet. Genom. 2016, 291, 1013–1033. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Zhang, J.; Zhou, Z.; Wang, L.; Liu, Y.; Liu, Y. ALDB: A domestic-animal long noncoding RNA database. PLoS ONE 2015, 10, e0124003. [Google Scholar] [CrossRef] [PubMed]
- Bohmdorfer, G.; Wierzbicki, A.T. Control of Chromatin Structure by Long Noncoding RNA. Trends Cell Biol. 2015, 25, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Lin, A.; Li, C.; Liang, K.; Wang, S.; Liu, Y.; Park, P.K.; Qin, L.; Wei, Y.; Hawke, D.H.; et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 2014, 159, 1110–1125. [Google Scholar] [CrossRef] [PubMed]
- Wan, G.; Hu, X.; Liu, Y.; Han, C.; Sood, A.K.; Calin, G.A.; Zhang, X.; Lu, X. A novel non-coding RNA lncRNA-JADE connects DNA damage signalling to histone H4 acetylation. EMBO J. 2013, 32, 2833–2847. [Google Scholar] [CrossRef] [PubMed]
- Gooding, A.J.; Zhang, B.; Jahanbani, F.K.; Gilmore, H.L.; Chang, J.C.; Valadkhan, S.; Schiemann, W.P. The lncRNA BORG drives breast cancer metastasis and disease recurrence. Sci. Rep. 2017, 7, 12698. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.A.; Chen, Z.; Park, J.T.; Wang, M.; Lanting, L.; Zhang, Q.; Bhatt, K.; Leung, A.; Wu, X.; Putta, S.; et al. Regulation of inflammatory phenotype in macrophages by a diabetes-induced long noncoding RNA. Diabetes 2014, 63, 4249–4261. [Google Scholar] [CrossRef] [PubMed]
- Shan, K.; Liu, C.; Liu, B.H.; Chen, X.; Dong, R.; Liu, X.; Zhang, Y.Y.; Liu, B.; Zhang, S.J.; Wang, J.J.; et al. Circular non-coding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation 2017. [Google Scholar] [CrossRef] [PubMed]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 2004, 431, 931–945. [Google Scholar] [CrossRef]
- Guttman, M.; Garber, M.; Levin, J.Z.; Donaghey, J.; Robinson, J.; Adiconis, X.; Fan, L.; Koziol, M.J.; Gnirke, A.; Nusbaum, C.; et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010, 28, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef] [PubMed]
- St Laurent, G.; Wahlestedt, C.; Kapranov, P. The Landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Andronescu, M.; Bereg, V.; Hoos, H.H.; Condon, A. RNA STRAND: The RNA secondary structure and statistical analysis database. BMC Bioinform. 2008, 9, 340. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Liu, X.; Liu, L.; Deng, H.; Zhang, J.; Xu, Q.; Cen, B.; Ji, A. Regulation of lncRNA expression. Cell. Mol. Biol. Lett. 2014, 19, 561–575. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.D.; Han, L.; Lee, H.; Zhuang, L.; Zhang, Y.; Baddour, J.; Nagrath, D.; Wood, C.G.; Gu, J.; Wu, X.; et al. Energy stress-induced lncRNA FILNC1 represses c-Myc-mediated energy metabolism and inhibits renal tumor development. Nat. Commun. 2017, 8, 783. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.-F.; Yang, L.; Chen, L.-L. The long noncoding RNA regulation at the MYC locus. Curr. Opin. Genet. Dev. 2015, 33, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Fenner, A. Kidney cancer: AR promotes RCC via lncRNA interaction. Nat. Rev. Urol. 2016, 13, 242. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Zhang, D.; Wu, W.; Wu, S.; Qian, J.; Hao, Y.; Yan, F.; Zhu, P.; Wu, J.; Huang, G.; et al. Mesenchymal stem cells promote hepatocarcinogenesis via lncRNA-MUF interaction with ANXA2 and miR-34a. Cancer Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, H.; Matsuda, Y.; Yamamoto, M.; Kamiya, S.; Ishiwata, T. Expression and role of long non-coding RNA H19 in carcinogenesis. Front. Biosci. 2018, 23, 614–625. [Google Scholar]
- Gabory, A.; Jammes, H.; Dandolo, L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. Bioessays 2010, 32, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.T.; He, J.; Liang, Q.; Ren, L.L.; Yan, T.T.; Yu, T.C.; Tang, J.Y.; Bao, Y.J.; Hu, Y.; Lin, Y.; et al. LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 2016, 6, 784–801. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Chen, T.; Li, G.; Yu, X.; Lu, Y.; Wang, H.; Teng, L. LncRNAs: Emerging biomarkers in gastric cancer. Future Oncol. 2015, 11, 2427–2441. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Somarowthu, S.; Legiewicz, M.; Chillon, I.; Marcia, M.; Liu, F.; Pyle, A.M. HOTAIR forms an intricate and modular secondary structure. Mol. Cell 2015, 58, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yang, L.; Zhong, T.; Mueller, M.; Men, Y.; Zhang, N.; Xie, J.; Giang, K.; Chung, H.; Sun, X.; et al. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat. Commun. 2015, 6, 10221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelley, R.L.; Kuroda, M.I. Noncoding RNA genes in dosage compensation and imprinting. Cell 2000, 103, 9–12. [Google Scholar] [CrossRef]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Nagano, T.; Mitchell, J.A.; Sanz, L.A.; Pauler, F.M.; Ferguson-Smith, A.C.; Feil, R.; Fraser, P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 2008, 322, 1717–1720. [Google Scholar] [CrossRef] [PubMed]
- Rogler, L.E.; Kosmyna, B.; Moskowitz, D.; Bebawee, R.; Rahimzadeh, J.; Kutchko, K.; Laederach, A.; Notarangelo, L.D.; Giliani, S.; Bouhassira, E.; et al. Small RNAs derived from lncRNA RNase MRP have gene-silencing activity relevant to human cartilage-hair hypoplasia. Hum. Mol. Genet. 2014, 23, 368–382. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yan, M.; Cao, Z.; Li, X.; Zhang, Y.; Shi, J.; Feng, G.H.; Peng, H.; Zhang, X.; Zhang, Y.; et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016, 351, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.C.; Di, C.; Hu, B.; Zhou, M.; Liu, Y.; Song, N.; Li, Y.; Umetsu, J.; Lu, Z.J. CLIPdb: A CLIP-seq database for protein-RNA interactions. BMC Genom. 2015, 16, 51. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed]
- Anko, M.L.; Neugebauer, K.M. RNA-protein interactions in vivo: Global gets specific. Trends Biochem. Sci. 2012, 37, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhu, Z.; Watabe, K.; Zhang, X.; Bai, C.; Xu, M.; Wu, F.; Mo, Y.Y. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013, 20, 1558–1568. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Lin, W.; Guo, M.; Zou, Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput. Biol. 2017, 13, e1005420. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Yao, J.; Liu, J.Y.; Li, X.M.; Wang, X.Q.; Li, Y.J.; Tao, Z.F.; Song, Y.C.; Chen, Q.; Jiang, Q. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ. Res. 2015, 116, 1143–1156. [Google Scholar] [CrossRef] [PubMed]
- Delli Ponti, R.; Marti, S.; Armaos, A.; Tartaglia, G.G. A high-throughput approach to profile RNA structure. Nucleic Acids Res. 2017, 45, e35. [Google Scholar] [CrossRef] [PubMed]
- Burgess, D.J. RNA. Detailed probing of RNA structure in vivo. Nat. Rev. Genet. 2015, 16, 255. [Google Scholar] [CrossRef] [PubMed]
- Spitale, R.C.; Flynn, R.A.; Zhang, Q.C.; Crisalli, P.; Lee, B.; Jung, J.W.; Kuchelmeister, H.Y.; Batista, P.J.; Torre, E.A.; Kool, E.T.; et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 2015, 519, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, Y.; Vigilante, A.; Darbo, E.; Zirra, A.; Militti, C.; D’Ambrogio, A.; Luscombe, N.M.; Ule, J. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 2015, 519, 491–494. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Talkish, J.; May, G.; Lin, Y.; Woolford, J.L., Jr.; McManus, C.J. Mod-seq: High-throughput sequencing for chemical probing of RNA structure. RNA 2014, 20, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Cruz, J.A.; Westhof, E. The dynamic landscapes of RNA architecture. Cell 2009, 136, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Lescoute, A.; Westhof, E. The interaction networks of structured RNAs. Nucleic Acids Res. 2006, 34, 6587–6604. [Google Scholar] [CrossRef] [PubMed]
- Dann, C.E.; Wakeman, C.A.; Sieling, C.L.; Baker, S.C.; Irnov, I.; Winkler, W.C. Structure and mechanism of a metal-sensing regulatory RNA. Cell 2007, 130, 878–892. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, P.C.; Ritchey, L.E.; Su, Z.; Assmann, S.M. Genome-wide analysis of RNA secondary structure. Annu. Rev. Genet. 2016, 50, 235–266. [Google Scholar] [CrossRef] [PubMed]
- Chiu, J.K.; Chen, Y.P. Pairwise RNA secondary structure alignment with conserved stem pattern. Bioinformatics 2015, 31, 3914–3921. [Google Scholar] [CrossRef] [PubMed]
- Incarnato, D.; Neri, F.; Anselmi, F.; Oliviero, S. RNA structure framework: Automated transcriptome-wide reconstruction of RNA secondary structures from high-throughput structure probing data. Bioinformatics 2016, 32, 459–461. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, J.H.; Spector, D.L. Long non-coding RNAs: Modulators of nuclear structure and function. Curr. Opin. Cell Biol. 2014, 26, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, B.; Hein, J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 2003, 31, 3423–3428. [Google Scholar] [CrossRef] [PubMed]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Alam, T.; Uludag, M.; Essack, M.; Salhi, A.; Ashoor, H.; Hanks, J.B.; Kapfer, C.; Mineta, K.; Gojobori, T.; Bajic, V.B. FARNA: Knowledgebase of inferred functions of non-coding RNA transcripts. Nucleic Acids Res. 2017, 45, 2838–2848. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z., Jr.; Hugenholtz, P.; Keller, K.; Brodie, E.L.; Larsen, N.; Piceno, Y.M.; Phan, R.; Andersen, G.L. NAST: A multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006, 34, W394–W399. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xiao, Y. Using 3dRNA for RNA 3-D structure prediction and evaluation. Curr. Protoc. Bioinform. 2017, 57, 5–9. [Google Scholar] [CrossRef]
- Lyngso, R.B.; Pedersen, C.N. RNA pseudoknot prediction in energy-based models. J. Comput. Biol. 2000, 7, 409–427. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Kertesz, M.; Spitale, R.C.; Segal, E.; Chang, H.Y. Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 2011, 12, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Xing, P.; Zou, Q. Detecting N6-methyladenosine sites from RNA transcriptomes using ensemble Support Vector Machines. Sci. Rep. 2017, 7, 40242. [Google Scholar] [CrossRef] [PubMed]
- Blythe, A.J.; Fox, A.H.; Bond, C.S. The ins and outs of lncRNA structure: How, why and what comes next? Biochim. Biophys. Acta 2016, 1859, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Rice, K.; Wang, Y.; Chen, W.; Zhong, Y.; Nakayama, Y.; Zhou, Y.; Klibanski, A. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: Isoform structure, expression, and functions. Endocrinology 2010, 151, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Novikova, I.V.; Hennelly, S.P.; Sanbonmatsu, K.Y. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012, 40, 5034–5051. [Google Scholar] [CrossRef] [PubMed]
- Uzilov, A.V.; Underwood, J.G. High-throughput nuclease probing of RNA structures using fragSeq. Methods Mol. Biol. 2016, 1490, 105–134. [Google Scholar] [CrossRef] [PubMed]
- Kertesz, M.; Wan, Y.; Mazor, E.; Rinn, J.L.; Nutter, R.C.; Chang, H.Y.; Segal, E. Genome-wide measurement of RNA secondary structure in yeast. Nature 2010, 467, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Ferre, F.; Colantoni, A.; Helmer-Citterich, M. Revealing protein-lncRNA interaction. Brief. Bioinform. 2016, 17, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.A.; Mattick, J.S. Structural and functional annotation of long noncoding RNAs. Methods Mol. Biol. 2017, 1526, 65–85. [Google Scholar] [CrossRef] [PubMed]
- Winkler, W.C.; Dann, C.E. RNA allostery glimpsed. Nat. Struct. Mol. Biol. 2006, 13, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Nie, F.; Wang, Y.; Zhang, Z.; Hou, J.; He, D.; Xie, M.; Xu, L.; De, W.; Wang, Z.; et al. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016, 76, 6299–6310. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.C.; Bloom, R.J.; Smolke, C.D. Engineering biological systems with synthetic RNA molecules. Mol. Cell 2011, 43, 915–926. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zheng, H.; Wang, C.; Lu, X.; Zhao, X.; Li, X. Insight into HOTAIR structural features and functions as landing pads for transcription regulation proteins. Biochem. Biophys. Res. Commun. 2017, 485, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Duszczyk, M.M.; Wutz, A.; Rybin, V.; Sattler, M. The Xist RNA A-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. RNA 2011, 17, 1973–1982. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhang, F.; Jiang, Y.; Hu, Y.; Gong, Z.; Liu, S.; Chen, X.; Jiang, Q.; Hao, J. Integrating genome-wide association studies and gene expression data highlights dysregulated multiple sclerosis risk pathways. Mult. Scler. 2017, 23, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Lister, N.; Shevchenko, G.; Walshe, J.L.; Groen, J.; Johnsson, P.; Vidarsdottir, L.; Grander, D.; Ataide, S.F.; Morris, K.V. The molecular dynamics of long noncoding RNA control of transcription in PTEN and its pseudogene. Proc. Natl. Acad. Sci. USA 2017, 114, 9942–9947. [Google Scholar] [CrossRef] [PubMed]
- Delebecque, C.J.; Lindner, A.B.; Silver, P.A.; Aldaye, F.A. Organization of intracellular reactions with rationally designed RNA assemblies. Science 2011, 333, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Yap, K.L.; Li, S.; Muñoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.-M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Koziol, M.J.; Rinn, J.L. RNA traffic control of chromatin complexes. Curr. Opin. Genet. Dev. 2010, 20, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Sleutels, F.; Zwart, R.; Barlow, D.P. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 2002, 415, 810–813. [Google Scholar] [CrossRef] [PubMed]
- Plath, K.; Fang, J.; Mlynarczyk-Evans, S.K.; Cao, R.; Worringer, K.A.; Wang, H.; de la Cruz, C.C.; Otte, A.P.; Panning, B.; Zhang, Y. Role of histone H3 lysine 27 methylation in X inactivation. Science 2003, 300, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Wutz, A.; Rasmussen, T.P.; Jaenisch, R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002, 30, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Brockdorff, N.; Kawano, S.; Tsutui, K.; Tsutui, K.; Nakagawa, S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell 2010, 19, 469–476. [Google Scholar] [CrossRef] [PubMed]


| LncRNA | Function | Reference (PMID) |
|---|---|---|
| Airn | Paternal specificity silencing | 10988110 |
| CCAT1-L | Influence the expression of the MYC locus | 27147598 |
| CCAT1-S | Influence the expression of the MYC locus | 26254776 |
| CDKN2B-HS1 | Suppression of the expression of cancer associated CDKN2A/CDKN2B genes | 20541999 |
| GAS5 | Suppress the expression of miR-21 | 20041488 |
| GClnc1 | Recruit WDR5 and KAT2A for specific histone modification | 26289363 |
| H19 | Maternal expression, rapidly down-regulated in most tissues after birth. | 28930564 |
| 20486113 | ||
| HOTAIR | Form multiple histone modifying complex involved in histone modification reactions | 20616235 |
| 25866246 | ||
| Linc-P21 | Play important role in the P53 pathway and activated by P53 for cell apoptosis | 20673990 |
| MALAT1 | Highly expressed in breast, colon, and prostate | 24297251 |
| Associated with many diseases | 22425269 | |
| MIAT | Regulate microvascular dysfunction | 25587098 |
| MEG3 | Suppress cancer | 20032057 |
| Rsx | Regulate the X chromosome silencing | 12649488 |
| SPA1 | Hormone receptor co-activator | 22362738 |
| XIST | Regulate the X chromosome silencing | 21947263 |
| 11780141 | ||
| 20833368 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Wang, L.; Ding, Y.; Lu, X.; Zhang, G.; Yang, J.; Zheng, H.; Wang, H.; Jiang, Y.; Xu, L. LncRNA Structural Characteristics in Epigenetic Regulation. Int. J. Mol. Sci. 2017, 18, 2659. https://doi.org/10.3390/ijms18122659
Wang C, Wang L, Ding Y, Lu X, Zhang G, Yang J, Zheng H, Wang H, Jiang Y, Xu L. LncRNA Structural Characteristics in Epigenetic Regulation. International Journal of Molecular Sciences. 2017; 18(12):2659. https://doi.org/10.3390/ijms18122659
Chicago/Turabian StyleWang, Chenguang, Lianzong Wang, Yu Ding, Xiaoyan Lu, Guosi Zhang, Jiaxin Yang, Hewei Zheng, Hong Wang, Yongshuai Jiang, and Liangde Xu. 2017. "LncRNA Structural Characteristics in Epigenetic Regulation" International Journal of Molecular Sciences 18, no. 12: 2659. https://doi.org/10.3390/ijms18122659
APA StyleWang, C., Wang, L., Ding, Y., Lu, X., Zhang, G., Yang, J., Zheng, H., Wang, H., Jiang, Y., & Xu, L. (2017). LncRNA Structural Characteristics in Epigenetic Regulation. International Journal of Molecular Sciences, 18(12), 2659. https://doi.org/10.3390/ijms18122659