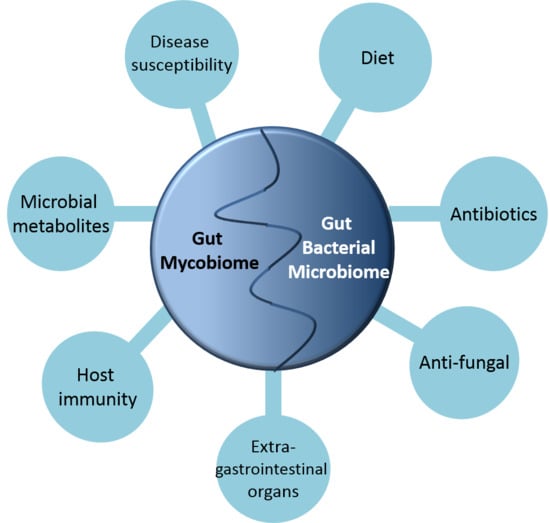
The Fungal Mycobiome and Its Interaction with Gut Bacteria in the Host
Abstract
:1. Introduction
2. Fungi That Are Found in the GI Tract
3. (Anti)-Fungal and (Anti)-Bacterial Interactions
4. The Gut Mycobiome and Disease Susceptibility
5. Dietary Influences and Obesity
6. The Mycobiome and Immunity
7. The Role of Extracellular Substances Produced by the Commensal Microbiota in the Gut in Preventing Pathogenicity and Maintaining the Myco-Microbiome Balance
8. Conclusions
Acknowledgments
Conflicts of Interest
References
- Dobell, C. The discovery of the intestinal protozoa of man. Proc. R. Soc. Med. 1920, 13, 1–15. [Google Scholar] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.; Knight, R.; Gordon, J.I. The human microbiome project: Exploring the microbial part of ourselves in a changing world. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Group, T.N.H.W.; Peterson, J.; Garges, S.; Giovanni, M.; McInnes, P.; Wang, L.; Schloss, J.A.; Bonazzi, V.; McEwen, J.E.; Wetterstrand, K.A.; et al. The NIH human microbiome project. Genome Res. 2009, 19, 2317–2323. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalog established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillevet, P.M.; Sikaroodi, M.; Torzilli, A.P. Analyzing salt-marsh fungal diversity: Comparing arisa fingerprinting with clone sequencing and pyrosequencing. Fungal Ecol. 2009, 2, 160–167. [Google Scholar] [CrossRef]
- Cui, L.; Morris, A.; Ghedin, E. The human mycobiome in health and disease. Genome Med. 2013, 5, 63. [Google Scholar] [CrossRef] [PubMed]
- Huffnagle, G.B.; Noverr, M.C. The emerging world of the fungal microbiome. Trends Microbiol. 2013, 21, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.; Roth, F.J.; Delgado, E.; Ahearn, D.G.; Kalser, M.H. Fungal flora of the normal human small and large intestine. N. Engl. J. Med. 1969, 280, 638–641. [Google Scholar] [CrossRef] [PubMed]
- Drgona, L.; Khachatryan, A.; Stephens, J.; Charbonneau, C.; Kantecki, M.; Haider, S.; Barnes, R. Clinical and economic burden of invasive fungal diseases in europe: Focus on pre-emptive and empirical treatment of aspergillus and candida species. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Chung, J.; Battaglia, T.; Henderson, N.; Jay, M.; Li, H.; D. Lieber, A.; Wu, F.; Perez-Perez, G.I.; Chen, Y.; et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 2016, 8, 343ra382. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakobsson, H.E.; Jernberg, C.; Andersson, A.F.; Sjölund-Karlsson, M.; Jansson, J.K.; Engstrand, L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS ONE 2010, 5, e9836. [Google Scholar] [CrossRef] [PubMed]
- Savage, D.C. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 1977, 31, 107–133. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.J. Regulation of candida albicans populations in the gastrointestinal tract: Mechanisms and significance in GI and systemic candidiasis. In Current Topics in Medical Mycology; McGinnis, M.R., Borgers, M., Eds.; Springer: New York, NY, USA, 1989; pp. 315–402. [Google Scholar]
- Morgan, X.C.; Huttenhower, C. Chapter 12: Human microbiome analysis. PLoS Comput. Biol. 2012, 8, e1002808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scanlan, P.D.; Marchesi, J.R. Micro-eukaryotic diversity of the human distal gut microbiota: Qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J. 2008, 2, 1183–1193. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Archaea and fungi of the human gut microbiome: Correlations with diet and bacterial residents. PLoS ONE 2013, 8, e66019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suhr, M.J.; Hallen-Adams, H.E. The human gut mycobiome: Pitfalls and potentials—A mycologist’s perspective. Mycologia 2015, 107, 1057–1073. [Google Scholar] [CrossRef] [PubMed]
- Mar Rodríguez, M.; Pérez, D.; Javier Chaves, F.; Esteve, E.; Marin-Garcia, P.; Xifra, G.; Vendrell, J.; Jové, M.; Pamplona, R.; Ricart, W.; et al. Obesity changes the human gut mycobiome. Sci. Rep. 2015, 5, 14600. [Google Scholar] [CrossRef] [PubMed]
- Von Rosenvinge, E.C.; Song, Y.; White, J.R.; Maddox, C.; Blanchard, T.; Fricke, W.F. Immune status, antibiotic medication and ph are associated with changes in the stomach fluid microbiota. ISME J. 2013, 7, 1354–1366. [Google Scholar] [CrossRef] [PubMed]
- Hamad, I.; Sokhna, C.; Raoult, D.; Bittar, F. Molecular detection of eukaryotes in a single human stool sample from senegal. PLoS ONE 2012, 7, e40888. [Google Scholar] [CrossRef] [PubMed]
- Ott, S.J.; Kühbacher, T.; Musfeldt, M.; Rosenstiel, P.; Hellmig, S.; Rehman, A.; Drews, O.; Weichert, W.; Timmis, K.N.; Schreiber, S. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand. J. Gastroenterol. 2008, 43, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Gaskins, H.R.; Croix, J.A.; Nakamura, N.; Nava, G.M. Impact of the intestinal microbiota on the development of mucosal defense. Clin. Infect. Dis. 2008, 46, S80–S86. [Google Scholar] [CrossRef] [PubMed]
- Weinstock, G.M. Genomic approaches to studying the human microbiota. Nature 2012, 489, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Kuczynski, J.; Lauber, C.L.; Walters, W.A.; Parfrey, L.W.; Clemente, J.C.; Gevers, D.; Knight, R. Experimental and analytical tools for studying the human microbiome. Nat. Rev. Genet. 2012, 13, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Di Bella, J.M.; Bao, Y.; Gloor, G.B.; Burton, J.P.; Reid, G. High throughput sequencing methods and analysis for microbiome research. J. Microbiol. Methods 2013, 95, 401–414. [Google Scholar] [CrossRef] [PubMed]
- McFrederick, Q.S.; Mueller, U.G.; James, R.R. Interactions between fungi and bacteria influence microbial community structure in the megachile rotundata larval gut. Proc. R. Soc. Lond. B 2014, 281. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, M.M.; Teixeira-Santos, R.; Silva, A.P.; Cruz, L.; Ricardo, E.; Pina-Vaz, C.; Rodrigues, A.G. The effect of antibacterial and non-antibacterial compounds alone or associated with antifugals upon fungi. Front. Microbiol. 2015, 6, 669. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.L.A.; Vieira-Silva, S.; Liston, A.; Raes, J. How informative is the mouse for human gut microbiota research? Dis. Models Mech. 2015, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Faith, J.J.; Rey, F.E.; O’Donnell, D.; Karlsson, M.; McNulty, N.P.; Kallstrom, G.; Goodman, A.L.; Gordon, J.I. Creating and characterizing communities of human gut microbes in gnotobiotic mice. ISME J. 2010, 4, 1094–1098. [Google Scholar] [CrossRef] [PubMed]
- Cole, G.T.; Seshan, K.R.; Lynn, K.T.; Franco, M. Gastrointestinal candidiasis: Histopathology of candida-host interactions in a murine model. Mycol. Res. 1993, 97, 385–408. [Google Scholar] [CrossRef]
- Van der Waaij, D.; Vries, J.M.B.-D.; der Wees, J.E.C.L.-V. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J. Hyg. 1971, 69, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.D. Influence of antibiotics or certain intestinal bacteria on orally administered candida albicans in germ-free and conventional mice. Infect. Immun. 1971, 4, 731–737. [Google Scholar] [PubMed]
- Samonis, G.; Gikas, A.; Anaissie, E.J.; Vrenzos, G.; Maraki, S.; Tselentis, Y.; Bodey, G.P. Prospective evaluation of effects of broad-spectrum antibiotics on gastrointestinal yeast colonization of humans. Antimicrob. Agents Chemother. 1993, 37, 51–53. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cobas, A.E.; Gosalbes, M.J.; Friedrichs, A.; Knecht, H.; Artacho, A.; Eismann, K.; Otto, W.; Rojo, D.; Bargiela, R.; von Bergen, M.; et al. Gut microbiota disturbance during antibiotic therapy: A multi-omic approach. Gut 2013, 62, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; Huse, S.; Sogin, M.L.; Relman, D.A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16s rRNA sequencing. PLoS Biol. 2008, 6, e280. [Google Scholar] [CrossRef] [PubMed]
- Erb Downward, J.R.; Falkowski, N.R.; Mason, K.L.; Muraglia, R.; Huffnagle, G.B. Modulation of post-antibiotic bacterial community reassembly and host response by candida albicans. Sci. Rep. 2013, 3, 2191. [Google Scholar] [CrossRef] [PubMed]
- Mason, K.L.; Erb Downward, J.R.; Mason, K.D.; Falkowski, N.R.; Eaton, K.A.; Kao, J.Y.; Young, V.B.; Huffnagle, G.B. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect. Immun. 2012, 80, 3371–3380. [Google Scholar] [CrossRef] [PubMed]
- Mason, K.L.; Erb Downward, J.R.; Falkowski, N.R.; Young, V.B.; Kao, J.Y.; Huffnagle, G.B. Interplay between the gastric bacterial microbiota and candida albicans during postantibiotic recolonization and gastritis. Infect. Immun. 2012, 80, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.R.; Graham, C.E.; Gagliano, B.C.; Lorenz, M.C.; Garsin, D.A. Enterococcus faecalis inhibits hyphal morphogenesis and virulence of candida albicans. Infect. Immun. 2013, 81, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Savage, D.C. Microbial interference between indigenous yeast and lactobacilli in the rodent stomach. J. Bacteriol. 1969, 98, 1278–1283. [Google Scholar] [PubMed]
- Garsin, D.A.; Lorenz, M.C. Candida albicans and enterococcus faecalis in the gut. Gut Microbes 2013, 4, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Zhang, F.; Yang, X.; Wu, N.; Jiang, W.; Li, X.; Li, X.; Liu, Y. Changes in the composition of intestinal fungi and their role in mice with dextran sulfate sodium-induced colitis. Sci. Rep. 2015, 5, 10416. [Google Scholar] [CrossRef] [PubMed]
- Kumamoto, C.A. Inflammation and gastrointestinal candida colonization. Curr. Opin. Microbiol. 2011, 14, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.L.; Lamas, B.; Liguori, G.; Hoffmann, T.W.; Sokol, H. Gut fungal microbiota: The Yin and Yang of inflammatory bowel disease. Inflamm. Bowel Dis. 2015, 21, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Sendid, B.; Hoarau, G.; Colombel, J.-F.; Poulain, D.; Ghannoum, M.A. Mycobiota in gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 77–87. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, H.; Main, J.; Pennington, C.R.; Parratt, D. Antibody to selected strains of saccharomyces cerevisiae (baker’s and brewer’s yeast) and candida albicans in crohn’s disease. Gut 1990, 31, 536–538. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, C.; Tang, C.; He, Q.; Li, N.; Li, J. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in crohn’s disease. J. Clin. Gastroenterol. 2014, 48, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Liguori, G.; Lamas, B.; Richard, M.L.; Brandi, G.; da Costa, G.; Hoffmann, T.W.; Di Simone, M.P.; Calabrese, C.; Poggioli, G.; Langella, P.; et al. Fungal dysbiosis in mucosa-associated microbiota of crohn’s disease patients. J. Crohn's Colitis 2015, 10, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.-P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal microbiota dysbiosis in IBD. Gut 2016. [Google Scholar] [CrossRef] [PubMed]
- Pineton de Chambrun, G.P.; Torres, J.; Darfeuille-Michaud, A.; Colombel, J.F. The role of anti(myco)bacterial interventions in the management of ibd: Is there evidence at all? Dig. Dis. 2012, 30, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, M.L.; Limon, J.J.; Bar, A.S.; Leal, C.A.; Gargus, M.; Tang, J.; Brown, J.; Funari, V.A.; Wang, H.L.; Crother, T.R.; et al. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe 2016, 19, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Krause, R.; Reisinger, E.C. Candida and antibiotic-associated diarrhoea. Clin. Microbiol. Infect. 2005, 11, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Levine, J.; Dykoski, R.K.; Janoff, E.N. Candida-associated diarrhea: A syndrome in search of credibility. Clin. Infect. Dis. 1995, 21, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Santelmann, H.; Howard, J.M. Yeast metabolic products, yeast antigens and yeasts as possible triggers for irritable bowel syndrome. Eur. J. Gastroenterol. Hepatol. 2005, 17, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Luan, C.; Xie, L.; Yang, X.; Miao, H.; Lv, N.; Zhang, R.; Xiao, X.; Hu, Y.; Liu, Y.; Wu, N.; et al. Dysbiosis of fungal microbiota in the intestinal mucosa of patients with colorectal adenomas. Sci. Rep. 2015, 5, 7980. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.X.; Jung, J.H.; Zhang, Z.F.; Kim, I.H. Effect of dietary β-glucan on growth performance, fecal microbial shedding and immunological responses after lipopolysaccharide challenge in weaned pigs. Anim. Feed Sci. Technol. 2013, 179, 85–92. [Google Scholar] [CrossRef]
- Ekenna, O.; Sherertz, R.J. Factors affecting colonization and dissemination of candida albicans from the gastrointestinal tract of mice. Infect. Immun. 1987, 55, 1558–1563. [Google Scholar] [PubMed]
- Moyes, D.L.; Naglik, J.R. Mucosal immunity and candida albicans infection. Clin. Dev. Immunol. 2011, 2011, 346307. [Google Scholar] [CrossRef] [PubMed]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.P.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M.; et al. Interactions between commensal fungi and the C-type lectin receptor dectin-1 influence colitis. Science 2012, 336, 1314–1317. [Google Scholar] [CrossRef] [PubMed]
- McAleer, J.P.; Nguyen, N.L.H.; Chen, K.; Kumar, P.; Ricks, D.M.; Binnie, M.; Armentrout, R.A.; Pociask, D.A.; Hein, A.; Yu, A.; et al. Pulmonary Th17 antifungal immunity is regulated by the gut microbiome. J. Immunol. 2016, 197, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Donia, M.S.; Fischbach, M.A. Small molecules from the human microbiota. Science 2015, 349, 1254766. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-J.; Hase, K. Gut microbiota-generated metabolites in animal health and disease. Nat. Chem. Biol. 2014, 10, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, M.; Mathur, R.; Chang, C. Gas and the microbiome. Curr. Gastroenterol. Rep. 2013, 15, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Midtvedt, T.; Gordon, J.I. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 2002, 22, 283–307. [Google Scholar] [CrossRef] [PubMed]
- Cottier, F.; Tan, A.S.M.; Chen, J.; Lum, J.; Zolezzi, F.; Poidinger, M.; Pavelka, N. The transcriptional stress response of candida albicans to weak organic acids. G3 Genes Genomes Genet. 2015, 5, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Shareck, J.; Belhumeur, P. Modulation of morphogenesis in candida albicans by various small molecules. Eukaryot. Cell 2011, 10, 1004–1012. [Google Scholar] [CrossRef] [PubMed]
- Swidergall, M.; Ernst, J.F. Interplay between candida albicans and the antimicrobial peptide armory. Eukaryot. Cell 2014, 13, 950–957. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Coughlin, L.A.; Neubauer, M.M.; Kim, J.; Kim, M.; Zhan, X.; Simms-Waldrip, T.R.; Xie, Y.; Hooper, L.V.; Koh, A.Y. Activation of HIF-1α and LL-37 by commensal bacteria inhibits candida albicans colonization. Nat. Med. 2015, 21, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.G.; Des Etages, S.G.; Snyder, M. Microbial synergy via an ethanol-triggered pathway. Mol. Cell. Biol. 2004, 24, 3874–3884. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sam, Q.H.; Chang, M.W.; Chai, L.Y.A. The Fungal Mycobiome and Its Interaction with Gut Bacteria in the Host. Int. J. Mol. Sci. 2017, 18, 330. https://doi.org/10.3390/ijms18020330
Sam QH, Chang MW, Chai LYA. The Fungal Mycobiome and Its Interaction with Gut Bacteria in the Host. International Journal of Molecular Sciences. 2017; 18(2):330. https://doi.org/10.3390/ijms18020330
Chicago/Turabian StyleSam, Qi Hui, Matthew Wook Chang, and Louis Yi Ann Chai. 2017. "The Fungal Mycobiome and Its Interaction with Gut Bacteria in the Host" International Journal of Molecular Sciences 18, no. 2: 330. https://doi.org/10.3390/ijms18020330
APA StyleSam, Q. H., Chang, M. W., & Chai, L. Y. A. (2017). The Fungal Mycobiome and Its Interaction with Gut Bacteria in the Host. International Journal of Molecular Sciences, 18(2), 330. https://doi.org/10.3390/ijms18020330