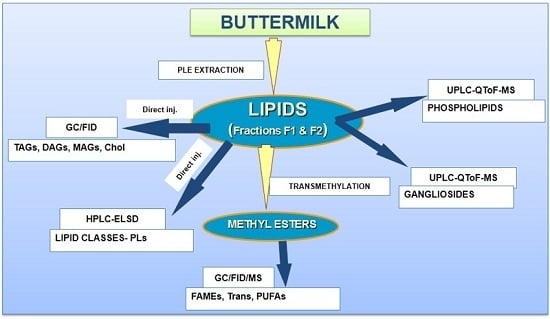
In-Depth Lipidomic Analysis of Molecular Species of Triacylglycerides, Diacylglycerides, Glycerophospholipids, and Sphingolipids of Buttermilk by GC-MS/FID, HPLC-ELSD, and UPLC-QToF-MS
Abstract
:1. Introduction
2. Results and Discussion
2.1. Fatty Acid Methyl Ester Analysis by GC-MS of Buttermilk and Isolated Fractions
2.2. Triacylgliceride Analysis of Buttermilk Fat and Isolated Fractions by GC-FID
2.3. Analysis of Buttermilk Fat and Isolated Lipid Fractions by UPLC/QToF MS
2.3.1. Identification and Quantification of Triacylglyceride and Diacylglyceride Molecular Species
2.3.2. Polar Lipid Molecular Species by UPLC-QTOF-MS
2.3.3. Glycosphingolipid Molecular Species by UPLC-QTOF-MS
3. Materials and Methods
3.1. Samples and Reagents
3.2. Total Fat Extraction by Pressurized Liquid Extraction System (PLE) and Lipid Fractionation by Flash Chromatography (FC)
3.3. Lipid Classes Composition by HPLC-ELSD
3.4. FA Determination and Quantification by GC-MS
3.5. Triacylglycerides and Cholesterol Determination
3.6. Phospholipid Molecular Species Determination by UPLC/QToF-MS
3.7. Triacylglyceride and Diacylglyceride Molecular Species Determination by UPLC/QToF-MS
3.8. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Danthine, S.; Blecker, C.; Paquot, M.; Innocente, N.; Deroanne, C. Évolution des connaissances sur la membrane du globule gras du lait: Synthèse bibliographique. Lait 2000, 80, 209–222. [Google Scholar] [CrossRef]
- Rodríguez-Alcalá, L.M.; Calvo, M.V.; Villar-Tajadura, M.A.; Castro-Gómez, M.d.P.; Holgado, F.; Juárez, M.; Fontecha, J. Lípidos bioactivos en productos lácteos. In Avances en la Investigación de la Alimentación Funcional; Fontecha, J., Ed.; Programa CYTED: Madrid, Spain, 2010. [Google Scholar]
- Castro-Gomez, P.; Garcia-Serrano, A.; Visioli, F.; Fontecha, J. Relevance of dietary glycerophospholipids and sphingolipids to human health. Prostaglandins Leukot. Essent. Fatty Acids 2015, 101, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Rombaut, R.; Camp, J.V.; Dewettinck, K. Phospho- and sphingolipid distribution during processing of milk, butter and whey. Int. J. Food Sci. Technol. 2006, 41, 435–443. [Google Scholar] [CrossRef]
- Kuchta, A.M.; Kelly, P.M.; Stanton, C.; Devery, R.A. Milk fat globule membrane—A source of polar lipids for colon health? A review. Int. J. Dairy Technol. 2012, 65, 315–333. [Google Scholar] [CrossRef]
- Conway, V.; Gauthier, S.F.; Pouliot, Y. Buttermilk: Much more than a source of milk phospholipids. Anim. Front. 2014, 4, 44–51. [Google Scholar] [CrossRef]
- Castro-Gómez, P.; Rodríguez-Alcalá, L.M.; Monteiro, K.M.; Ruiz, A.L.T.G.; Carvalho, J.E.; Fontecha, J. Antiproliferative activity of buttermilk lipid fractions isolated using food grade and non-food grade solvents on human cancer cell lines. Food Chem. 2016, 212, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Kullenberg, D.; Taylor, L.A.; Schneider, M.; Massing, U. Health effects of dietary phospholipids. Lipids Health Dis. 2012, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- El-Loly, M.M. Composition, properties and nutritional aspects of milk fat globule membrane—A Review. Pol. J. Food Nutr. Sci. 2011, 61, 7–32. [Google Scholar] [CrossRef]
- Caboni, M.; Menotta, S.; Lercker, G. Separation and analysis of phospholipids in different foods with a light-scattering detector. J. Am. Oil Chem. Soc. 1996, 73, 1561–1566. [Google Scholar] [CrossRef]
- Fong, B.Y.; Norris, C.S.; MacGibbon, A.K.H. Protein and lipid composition of bovine milk-fat-globule membrane. Int. Dairy J. 2007, 17, 275–288. [Google Scholar] [CrossRef]
- Costa, M.R.; Elias-Argote, X.E.; Jiménez-Flores, R.; Gigante, M.L. Use of ultrafiltration and supercritical fluid extraction to obtain a whey buttermilk powder enriched in milk fat globule membrane phospholipids. Int. Dairy J. 2010, 20, 598–602. [Google Scholar] [CrossRef]
- Konrad, G.; Kleinschmidt, T.; Lorenz, C. Ultrafiltration of whey buttermilk to obtain a phospholipid concentrate. Int. Dairy J. 2013, 30, 39–44. [Google Scholar] [CrossRef]
- Castro-Gómez, P.; Rodriguez-Alcalá, L.M.; Calvo, M.V.; Holgado, F.; Carvalho, J.E.; Fontecha, J. Antiproliferative activity of buttermilk milk fat globule membrane and isolated lipid fractions on ten human cancer cell lines. Ann. Nutr. Metab. 2013, 63, 1623. [Google Scholar]
- Castro-Gomez, M.P.; Rodriguez-Alcala, L.M.; Calvo, M.V.; Romero, J.; Mendiola, J.A.; Ibanez, E.; Fontecha, J. Total milk fat extraction and quantification of polar and neutral lipids of cow, goat, and ewe milk by using a pressurized liquid system and chromatographic techniques. J. Dairy Sci. 2014, 97, 6719–6728. [Google Scholar] [CrossRef] [PubMed]
- Castro-Gómez, P.; Fontecha, J.; Rodríguez-Alcalá, L.M. A high-performance direct transmethylation method for total fatty acids assessment in biological and foodstuff samples. Talanta 2014, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Alcalá, L.M.; Fontecha, J. Hot topic: Fatty acid and conjugated linoleic acid (CLA) isomer composition of commercial CLA-fortified dairy products: Evaluation after processing and storage. J. Dairy Sci. 2007, 90, 2083–2090. [Google Scholar] [CrossRef] [PubMed]
- Sokol, E.; Ulven, T.; Færgeman, N.J.; Ejsing, C.S. Comprehensive and quantitative profiling of lipid species in human milk, cow milk and a phospholipid-enriched milk formula by GC and MS/MSALL. Eur. J. Lipid Sci. Technol. 2015, 117, 751–759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristensen, D.; Hedegaard, R.V.; Nielsen, J.H.; Skibsted, L.H. Oxidative stability of buttermilk as influenced by the fatty acid composition of cows’ milk manipulated by diet. J. Dairy Res. 2004, 71, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Fontecha, J.; Rodriguez-Alcala, L.M.; Calvo, M.V.; Juarez, M. Bioactive Milk Lipids. Curr. Nutr. Food Sci. 2011, 7, 155–159. [Google Scholar] [CrossRef]
- Smith, L.M.; Lowry, R.R. Fatty Acid Composition of the Phospholipids and Other Lipids in Milk. J. Dairy Sci. 1962, 45, 581–588. [Google Scholar] [CrossRef]
- Castro-Gómez, M.; Holgado, F.; Rodríguez-Alcalá, L.; Montero, O.; Fontecha, J. Comprehensive Study of the Lipid Classes of Krill Oil by Fractionation and Identification of Triacylglycerols, Diacylglycerols, and Phospholipid Molecular Species by Using UPLC/QToF-MS. Food Anal. Methods 2015, 10, 2568–2580. [Google Scholar]
- Araujo, P.; Zhu, H.; Breivik, J.F.; Hjelle, J.I.; Zeng, Y. Determination and structural elucidation of triacylglycerols in krill oil by chromatographic techniques. Lipids 2014, 49, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Szabo, A.; Mézes, M.; Hancz, C.; Molnár, T.; Varga, D.; Romvári, R.; Fébel, H. Incorporation dynamics of dietary vegetable oil fatty acids into the triacylglycerols and phospholipids of tilapia (Oreochromis niloticus) tissues (fillet, liver, visceral fat and gonads). Aquacult. Nutr. 2011, 17, 132–147. [Google Scholar] [CrossRef]
- Ali-Nehari, A.; Chun, B.S. Characterization of purified phospholipids from krill (Euphausia superba) residues deoiled by supercritical carbon dioxide. Korean J. Chem. Eng. 2012, 29, 918–924. [Google Scholar] [CrossRef]
- Wang, T.; Hammond, E.; Fehr, W. Phospholipid fatty acid composition and stereospecific distribution of soybeans with a wide range of fatty acid composition. J. Am. Oil Chem. Soc. 1997, 74, 1587–1594. [Google Scholar] [CrossRef]
- Sánchez-Juanes, F.; Alonso, J.M.; Zancada, L.; Hueso, P. Distribution and fatty acid content of phospholipids from bovine milk and bovine milk fat globule membranes. Int. Dairy J. 2009, 19, 273–278. [Google Scholar]
- Zancada, L.; Pérez-Díez, F.; Sánchez-Juanes, F.; Alonso, J.M.; García-Pardo, L.A.; Hueso, P. Phospholipid classes and fatty acid composition of ewe’s and goat’s milk. Grasas Y Aceites 2013, 64, 304–310. [Google Scholar]
- Lopez, C.; Briard-Bion, V.; Menard, O.; Rousseau, F.; Pradel, P.; Besle, J.M. Phospholipid, sphingolipid, and fatty acid compositions of the milk fat globule membrane are modified by diet. J. Agric. Food Chem. 2008, 56, 5226–5236. [Google Scholar] [CrossRef] [PubMed]
- Fontecha, J.; Mayo, I.; Toledano, G.; Juarez, M. Triacylglycerol composition of protected designation of origin cheeses during ripening. Authenticity of milk fat. J. Dairy Sci. 2006, 89, 882–887. [Google Scholar] [CrossRef]
- Fraga, M.J.; Fontecha, J.; Lozada, L.; Juarez, M. Silver ion adsorption thin layer chromatography and capillary gas chromatography in the study of the composition of milk fat triglycerides. J. Agric. Food Chem 1998, 46, 1836–1843. [Google Scholar] [CrossRef]
- Gille, D. The health aspects of buttermilk components. A review. Technical-scientific information. ALP Sci. 2011, 540, 1–22. [Google Scholar]
- Calvano, C.D.; Jensen, O.N.; Zambonin, C.G. Selective extraction of phospholipids from dairy products by micro-solid phase extraction based on titanium dioxide microcolumns followed by MALDI-TOF-MS analysis. Anal. Bioanal. Chem. 2009, 394, 1453–1461. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, K.; Eaton, S. Mitochondrial β-oxidation. Eur. J. Biochem. 2004, 271, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Shindou, H.; Hishikawa, D.; Harayama, T.; Yuki, K.; Shimizu, T. Recent progress on acyl CoA: Lysophospholipid acyltransferase research. J. Lipid Res. 2009, 50, S46–S51. [Google Scholar] [CrossRef] [PubMed]
- Goñi, F.M.; Alonso, A. Structure and functional properties of diacylglycerols in membranes1. Prog. Lipid Res. 1999, 38, 1–48. [Google Scholar] [CrossRef]
- Becker, K.P.; Hannun, Y.A. Chapter 2—Diacylglycerols. In Bioactive Lipids; Nicolaou, A., Kokotos, G., Eds.; Woodhead Publishing: Cambridge, UK, 2004; pp. 37–61. [Google Scholar]
- Fagan, P.; Wijesundera, C.; Watkins, P. Determination of mono- and di-acylglycerols in milk lipids. J. Chromatogr. A 2004, 1054, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Gallier, S.; Gragson, D.; Cabral, C.; Jiménez-Flores, R.; Everett, D.W. Composition and fatty acid distribution of bovine milk phospholipids from processed milk products. J. Agric. Food Chem. 2010, 58, 10503–10511. [Google Scholar] [CrossRef] [PubMed]
- Calvano, C.D.; de Ceglie, C.; Aresta, A.; Facchini, L.A.; Zambonin, C.G. MALDI-TOF mass spectrometric determination of intact phospholipids as markers of illegal bovine milk adulteration of high-quality milk. Anal. Bioanal. Chem. 2013, 405, 1641–1649. [Google Scholar] [CrossRef] [PubMed]
- Bitman, J.; Wood, D.L. Changes in milk fat phospholipids during lactation. J. Dairy Sci. 1990, 73, 1208–1216. [Google Scholar] [CrossRef]
- Lopez, C.; Briard-Bion, V.; Ménard, O. Polar lipids, sphingomyelin and long-chain unsaturated fatty acids from the milk fat globule membrane are increased in milks produced by cows fed fresh pasture based diet during spring. Food Res. Int. 2014, 59–68. [Google Scholar] [CrossRef]
- Sørensen, L.K. A liquid chromatography/tandem mass spectrometric approach for the determination of gangliosides GD3 and GM3 in bovine milk and infant formulae. Rapid Commun. Mass Spectrom. 2006, 20, 3625–3633. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; An, H.J.; Lerno, L.A., Jr.; German, J.B.; Lebrilla, C.B. Rapid profiling of bovine and human milk gangliosides by matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry. Int. J. Mass Spectrom. 2011, 305, 138–150. [Google Scholar] [CrossRef] [PubMed]
- García, A.D.; Chavez, J.L.; Mechref, Y. Rapid and sensitive LC-ESI-MS of gangliosides. J. Chromatogr. B 2014, 947, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Montero, O.; Velasco, M.; Rodríguez-Lázaro, D.; Hernández, M. Analysis of cheese small molecules by UPLC-QToF-MS and multivariate statistical methods using several extraction procedures. Food Anal. Methods 2013, 6, 1497–1507. [Google Scholar] [CrossRef]
- Laegreid, A.; Otnaes, A.B.K.; Fuglesang, J. Human and bovine milk: Comparison of ganglioside composition and Enterotoxin-inhibitory activity. Pediatric Res. 1986, 20, 416–421. [Google Scholar] [CrossRef] [PubMed]
- Kolter, T. Ganglioside Biochemistry. Int. Sch. Res. Netw. Biochem. 2012. [Google Scholar] [CrossRef] [PubMed]




| Fatty Acid | BM | F1 | F2 |
|---|---|---|---|
| C4:0 | 4.33 a ± 0.19 | 4.44 a ± 0.22 | 4.65 a ± 0.07 |
| C6:0 | 2.39 a ± 0.15 | 2.25 a ± 0.12 | 2.55 a ± 0.02 |
| C8:0 | 1.23 a ± 0.07 | 1.48 a ± 0.06 | 1.21 a ± 0.01 |
| C10:0 | 2.70 a ± 0.10 | 2.73 a ± 0.01 | 2.63 a ± 0.06 |
| C10:1 | 0.22 a ± 0.02 | 0.28 a ± 0.01 | 0.21 a ± 0.01 |
| C12:0 | 3.11 a ± 0.12 | 3.36 a ± 0.01 | 3.08 a ± 0.04 |
| C14:0 | 11.69 a ± 0.31 | 12.55 a ± 0.08 | 12.14 a ± 0.04 |
| C15:0 anteiso | 0.22 a ± 0.01 | 0.20 a ± 0.01 | 0.22 a ± 0.01 |
| C15:0 iso | 0.42 a ± 0.01 | 0.35 b ± 0.01 | 0.43 a ± 0.01 |
| C14:1 | 0.54 a ± 0.02 | 0.27 b ± 0.01 | 0.36 c ± 0.01 |
| C15:0 | 1.19 a ± 0.02 | 1.32 b ± 0.01 | 1.20 a ± 0.01 |
| C16:0 iso | 0.28 a ± 0.01 | 0.25 b ± 0.01 | 0.30 a ± 0.01 |
| C16:0 | 35.39 a ± 0.05 | 34.47 a ± 0.34 | 39.96 b ± 0.09 |
| C17:0 anteiso | 0.38 a ± 0.03 | 0.28 b ± 0.02 | 0.41 a ± 0.01 |
| C16:1 | 1.42 a ± 0.03 | 1.19 b ± 0.01 | 1.07 b ± 0.01 |
| C17:0 | 0.63 a,b ± 0.03 | 0.51 a ± 0.01 | 0.73 b ± 0.03 |
| C18:0 | 11.23 a,b ± 0.36 | 9.89 a ± 0.07 | 13.01 b ± 0.08 |
| C18:1 t6–8 | 0.18 a ± 0.01 | 0.10 b ± 0.01 | 0.18 a ± 0.01 |
| C18:1 t9 | 0.22 a ± 0.01 | 0.20 a ± 0.01 | 0.18 a ± 0.01 |
| C18:1 t10 | 0.29 a ± 0.04 | 0.23 b ± 0.01 | 0.31 a ± 0.01 |
| C18:1 t11 | 1.02 a ± 0.02 | 0.82 b ± 0.01 | 1.10 c ± 0.01 |
| C18:1 t12 | 0.30 a ± 0.01 | 0.19 b ± 0.01 | 0.31 a ± 0.01 |
| C18:1 c9 | 17.34 a ± 0.29 | 18.51 a ± 0.06 | 10.73 b ± 0.01 |
| C18:1 t15 | 0.31 a ± 0.03 | 0.31 a ± 0.04 | 0.34 a ± 0.03 |
| C18:1 c11 | 0.59 a ± 0.04 | 0.59 a ± 0.01 | 0.34 b ± 0.03 |
| C18:1 c12 | 0.25 a ± 0.01 | 0.20 b ± 0.01 | 0.15 c ± 0.01 |
| C18:1 t14 + 16 | 0.21 a ± 0.02 | 0.11 b ± 0.01 | 0.23 a ± 0.01 |
| Total trans C18:1 | 2.54 a ± 0.13 | 1.84 b ± 0.06 | 2.42 a ± 0.07 |
| C18:2 c9,c12 | 0.60 a ± 0.02 | 1.75 b ± 0.03 | 0.41 c ± 0.02 |
| C18:2 c9,t11 | 0.03 a ± 0.01 | 0.09 b ± 0.01 | 0.15 c ± 0.01 |
| C20:0 | 0.10 a ± 0.02 | 0.07 a ± 0.01 | 0.11 a ± 0.02 |
| ∑ SFA | 75.96 a ± 0.55 a | 74.78 a ± 0.04 | 83.52 b ± 0.10 |
| ∑ MUFA | 23.31 a ± 0.43 a | 23.21 a ± 0.06 | 15.68 b ± 0.11 |
| ∑ PUFA | 0.74 a ± 0.11a | 2.01 b ± 0.02 | 0.79 a ± 0.02 |
| ∑ cis C18:1 | 18.23 a ± 0.36 a | 19.35 a ± 0.07 | 11.26 b ± 0.06 |
| ∑ trans C18:1 | 2.54 a ± 0.13 a | 1.84 b ± 0.06 | 2.42 a ± 0.07 |
| TAG Group (%) | BM | F2 | p |
|---|---|---|---|
| TAG CN28 | 1.03 ± 0.04 | <0.1 | 0.001 |
| TAG CN30 | 1.65 ± 0.10 | <0.1 | 0.001 |
| TAG CN32 | 4.29 ± 0.21 | 1.40 ± 0.24 | 0.000 |
| TAG CN34 | 8.46 ± 3.20 | 10.90 ± 0.81 | 0.046 |
| TAG CN36 | 10.54 ± 0.17 | 19.04 ± 0.98 | 0.004 |
| TAG CN38 | 18.30 ± 0.11 | 16.80 ± 0.45 | 0.051 |
| TAG CN40 | 12.53 ± 0.27 | 12.62 ± 0.62 | 0.830 |
| TAG CN42 | 8.17 ± 0.41 | 9.17 ± 0.37 | 0.957 |
| TAG CN44 | 7.61 ± 0.66 | 7.93 ± 0.30 | 0.494 |
| TAG CN46 | 6.48 ± 0.43 | 6.49 ± 0.63 | 0.984 |
| TAG CN48 | 7.69 ± 1.10 | 6.09 ± 0.43 | 0.093 |
| TAG CN50 | 7.59 ± 0.67 | 5.99 ± 0.70 | 0.871 |
| TAG CN52 | 5.65 ± 0.86 | 3.58 ± 1.08 | 0.742 |
| TAG CN54 | <0.1 | <0.1 | - |
| Time (min) | Exact Mass | Content (%) | CN/DB | Molecular Species |
|---|---|---|---|---|
| 0.93 | 470.3616 | 0.18 | 24:0 | TAG(8:0/8:0/8:0) |
| 0.97 | 578.4507 | 0.15 | 32:2 | TAG(14:1/14:1/4:0) |
| 0.98 | 496.3795 | 0.08 | 26:1 | TAG(14:1/8:0/4:0) |
| 1.03 | 498.3913 | 0.64 | 26:0 | TAG(14:0/8:0/4:0) |
| 1.07 | 524.4069 | 0.29 | 28:1 | TAG(16:1/8:0/4:0) |
| 1.13 | 526.4207 | 1.02 | 28:0 | TAG(16:0/8:0/4:0) |
| 1.45 | 608.4994 | 1.49 | 34:1 | TAG(16:1/14:0/4:0) |
| 1.68 | 636.5303 | 2.41 | 36:1 | TAG(14:0/14:1/8:0) |
| 1.72 | 610.5145 | 5.58 | 34:0 | TAG(16:0/12:0/6:0) |
| 1.73 | 662.5461 | 0.84 | 38:2 | TAG(16:1/16:1/6:0) |
| 1.96 | 690.5734 | 1.06 | 40:2 | TAG(14:1/14:1/12:0) |
| 2.03 | 638.5447 | 7.60 | 36:0 | TAG(12:0/12:0/12:0) |
| 2.40 | 666.5769 | 8.81 | 38:0 | TAG(16:0/16:0/6:0) |
| 2.67 | 772.6514 | 0.21 | 48:3 | TAG(16:1/16:1/16:1) |
| 2.67 | 720.6229 | 3.36 | 42:1 | TAG(18:1/16:0/8:0) |
| 2.70 | 694.6105 | 7.59 | 40:0 | TAG(16:0/16:0/8:0) |
| 2.95 | 734.6395 | 0.46 | 43:1 | TAG(14:0/14:1/15:0) |
| 3.15 | 774.6678 | 1.00 | 46:2 | TAG(16:1/16:0/14:1) |
| 3.18 | 748.6538 | 4.51 | 44:1 | TAG(16:0/14:0/14:1) |
| 3.25 | 722.6372 | 0.71 | 42:0 | TAG(16:0/14:0/12:0) |
| 3.53 | 788.6851 | 0.01 | 47:2 | TAG(14:1/15:0/18:1) |
| 3.84 | 802.6989 | 1.50 | 48:2 | TAG(16:0/16:1/16:1) |
| 4.71 | 830.7292 | 2.97 | 50:2 | TAG(16:1/18:1/16:0) |
| 3.87 | 776.6866 | 4.39 | 46:1 | TAG(18:1/14:0/14:0) |
| 4.30 | 790.7012 | 1.67 | 47:1 | TAG(18:1/14:0/15:0) |
| 4.37 | 764.6894 | 1.55 | 45:0 | TAG(16:0/15:0/14:0) |
| 4.71 | 856.7448 | 1.37 | 52:3 | TAG(16:0/18:1/18:2) |
| 4.77 | 804.7145 | 6.17 | 48:1 | TAG(14:0/18:1/16:0) |
| 4.87 | 778.6984 | 3.13 | 46:0 | TAG(14:0/14:0/18:0) |
| 5.12 | 818.7254 | 1.79 | 49:1 | TAG(15:0/16:0/18:1) |
| 5.19 | 792.7154 | 0.75 | 47:0 | TAG(16:0/14:0/17:0) |
| 5.33 | 818.7254 | 0.69 | 49:1 | TAG(16:0/18:1/15:0) |
| 5.43 | 792.7154 | 0.80 | 47:0 | TAG(16:0/16:0/15:0) |
| 5.94 | 832.7463 | 7.04 | 50:1 | TAG(16:0/16:0/18:1) |
| 6.05 | 806.7285 | 3.19 | 48:0 | TAG(16:0/16:0/16:0) |
| 6.19 | 828.7155 | 0.28 | 50:3 | TAG(18:2/18:1/14:0) |
| 6.22 | 884.7746 | 1.13 | 54:3 | TAG(18:1/18:1/18:1) |
| 6.37 | 872.7741 | 0.72 | 53:2 | TAG(18:1/18:1/17:0) |
| 6.40 | 846.7602 | 1.41 | 51:1 | TAG(16:0/17:0/18:1) |
| 6.46 | 820.7466 | 1.13 | 49:0 | TAG(16:0/16:0/17:0) |
| 6.81 | 820.7466 | 0.89 | 49:0 | TAG(16:0/15:0/18:0) |
| 7.39 | 834.7604 | 0.19 | 50:0 | TAG(16:0/16:0/18:0) |
| 7.50 | 886.7917 | 2.60 | 54:2 | TAG(18:0/18:1/18:1) |
| 7.64 | 860.7755 | 5.50 | 52:1 | TAG(16:0/18:0/18:1) |
| Positive Mode | Negative Mode | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Time (min) | Exact Mass | Content (%) | CN/DB | Molecular Species | Time (min) | Exact Mass | Content (%) | CN/DB | Molecular Species |
| 3.75 | 496.3418 | 0.31 | 16:0 | Lyso-PC(16:0) | 3.35 | 512.30 | 2.10 | 14:0 | Lyso-PC(14:0) |
| 3.93 | 522.355 | 0.24 | 18:1 | Lyso-PC(18:1) | 4.08 | 540.33 | 26.29 | 16:0 | Lyso-PC(16:0) |
| 4.95 | 524.368 | 0.20 | 18:0 | Lyso-PC(18:0) | 4.28 | 566.35 | 6.72 | 18:1 | Lyso-PC(18:1) |
| 6.53 | 734.5698 | 13.09 | 32:0 | PC(16:0/16:0) | 6.71 | 802.56 | 9.03 | 34:2 | PC(16:0/18:2) |
| 6.55/8.03 | 756.5527 | 1.21 | 34:3 | PC(16:1/18:2) | 6.8 | 828.58 | 1.99 | 36:3 | PC(18:1/18:2) |
| 6.66 | 760.5854 | 29.61 | 34:1 | PC(16:0/18:1) | 7.45 | 812.58 | 6.23 | 36:4/36:3 | PC(O-18:2/18:2) |
| 6.82 | 786.6024 | 9.38 | 36:2 | PC(18:1/18:1) | /(P-18:1/18:2) | ||||
| 7.03 | 786.6024 | 4.71 | 36:3 | PC(18:0/18:2) | 8.27 | 778.56 | 14.12 | 32:2 | PC(16:0/16:0) |
| 7.56 | 678.5092 | 1.94 | 28:0 | PC(14:0/14:0), (12:0/16:0) | 8.39 | 804.57 | 23.29 | 34:1 | PC(16:0/18:1) |
| 7.75 | 730.5361 | 0.65 | 32:2 | PC(16:1/16:1) | 8.49 | 722.50 | 1.55 | 28:0 | PC(12:0/16:0), |
| 8.02/8.5 | 782.5671 | 1.72 | 36:4 | PC(18:2/18:2) | (14:0/14:0) | ||||
| 8.46 | 706.5352 | 11.72 | 30:0 | PC(14:0/16:0), (12:0/18:0) | 8.52 | 830.59 | 7.13 | 36:2 | PC(18:1/18:1) |
| 8.55 | 732.5501 | 4.65 | 32:1 | PC(16:0/16:1) | 9.43 | 814.60 | 1.56 | 36:3/36:2 | PC(O-18:1/18:2) |
| 8.6 | 762.6018 | 2.56 | 34:0 | PC(16:0/18:0) | /(P-18:0/18:2) | ||||
| 8.76 | 758.5701 | 8.82 | 34:2 | PC(16:0/18:2) | |||||
| 8.79 | 788.6178 | 4.95 | 36:1 | PC(18:0/18:1) | |||||
| 8.84 | 784.5816 | 4.24 | 36:3 | PC(18:1/18:2) | |||||
| 8.84 | 784.5816 | 4.24 | 36:3 | PC(18:1/18:2) | |||||
| 5.58 | 705.588 | 5.16 | 34:0 | SM(d18:0/16:0) | 6.37 | 747.56 | 34.67 | 34:1 | SM(d18:1/16:0) |
| 6.16 | 759.637 | 0.81 | 38:1 | SM(d18:1/20:0) | 7.47 | 749.58 | 29.64 | 34:0 | SM(d18:0/16:0) |
| 7.69 | 675.5415 | 14.90 | 32:1 | SM(d16:1/16:0) | 8.66 | 719.53 | 24.49 | 32:1 | SM(d16:1/16:0) |
| 8.19 | 677.5576 | 6.53 | 32:0 | SM(d18:0/14:0) | 9.32 | 721.55 | 11.19 | 32:0 | SM(d18:0/14:0) |
| 8.18 | 689.5562 | 7.92 | 33:1 | SM(d18:1/15:0) | |||||
| 8.58/8.77 | 703.5729 | 64.69 | 34:1 | SM(d18:1/16:0) | |||||
| Positive Mode | Negative Mode | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Time (min) | Exact Mass | Content (%) | CN/DB | Molecular Species | Time (min) | Exact Mass | Content (%) | CN/DB | Molecular Species |
| 3.99 | 480.3053 | 1.13 | 18:1 | Lyso-PE(18:1) | 3.53 | 476.2809 | 0.81 | 18:2 | Lyso-PE(18:2) |
| 7.07 | 718.5381 | 2.24 | 34:1 | PE(16:0/18:1), (16:1/18:0) | 4.09 | 478.2934 | 1.12 | 16:0 | Lyso-PE(16:0) |
| 7.22/7.45/8.21 | 744.5569 | 9.72 | 36:2 | PE(18:1/18:1), (18:0/18:2) | 4.3 | 478.2934 | 1.04 | 18:1 | Lyso-PE(18:1) |
| 7.99 | 692.5201 | 9.46 | 32:0 | PE(16:0/16:0), (20:0/12:0), (18:0/14:0), (17:0/15:0) | 6.61 | 688.4931 | 2.72 | 32:2 | PE(14:0/18:2) |
| 8.86/9.1 | 720.5502 | 56.58 | 34:0 | PE(20:0/14:0), (18:0/16:0), (17:0/17:0) | 7.19 | 740.5279 | 26.89 | 36:3 | PE(18:1/18:2) |
| 8.95 | 746.5662 | 8.70 | 36:1 | PE(18:0/18:1), (20:0/16:1) | 7.06 | 714.5073 | 9.39 | 34:2 | PE(16:1/18:1), (16:0/18:2) |
| 9.06 | 716.5188 | 12.14 | 34:2 | PE(16:1/18:1), (18:2/16:0) | 8.87 | 716.5193 | 15.89 | 34:1 | PE(16:0/18:1), (16:1/18:0) |
| 9.06 | 742.5395 | 42.13 | 36:2 | PE(18:1/18:1), (18:0/18:2) | |||||
| 6.7 | 790.5614 | 39.37 | 36:1 | PS(18:1/18:0), (16:1/20:0) | 7.45 | 786.5295 | 80.91 | 36:2 | PS(18:1/18:1), (18:2/18:2) |
| 6.99 | 792.5767 | 60.63 | 36:0 | PS(16:0/20:0), (18:0/18:0) | 9.39 | 788.5384 | 19.09 | 36:1 | PS(18:0/18:1), (16:1/20:0) |
| 8.26 | 854.5759 | 19.69 | 34:1 | PI(16:0/18:1), (16:1/18:0) | 7.76 | 863.5589 | 100.00 | 36:1 | PI(18:0/18:1) |
| 8.33/8.46 | 880.5894 | 80.31 | 36:2 | PI(18:1/18:1), (18:0/18:2) | |||||
| No. | m/z | Ion | Exact Mass (Mexp − MTh) | Elemental Composition | Glycosphingolipid Class | Tentative Ceramide |
|---|---|---|---|---|---|---|
| 1 | 1013.75 | [M + 2H]+2 | 2025.11 (+0.37) | C94H168N4O42 | Globo/Lacto/Neolacto | -Cer(d18:1/22:0) |
| 2 | 1187.87 | [M + 2H]+2 | 2373.23 (+0.49) | C107H188N6O51 | Acidic | -Cer(d18:1/22:0) |
| 3 | 984.72 | [M + 2H]+2 | 1967.07 (+0.35) | C91H162N4O41 | Acidic | -Cer(d18:1/22:0) |
| 4 | 797.92 | [M + 3H]+3 | 2390.25 (+0.48) | C108H191N5O52 | Lacto/Neolacto | -Cer(d18:1/22:0) |
| 5 | 875.31 | [M + 3H]+3 | 2623.30 (−0.39) | C116H202N6O59 | Acidic (4 isomers) | -Cer(d18:1/22:0) |
| 6 | 855.97 | [M + 3H]+3 | 2565.29 (−0.43) | C114H200N6O57 | Lacto/Neolacto | -Cer(d18:1/20:0) |
| 2565.32 (−0.46) | C116H204N4O57 | Neolacto (2 isomers) | -Cer(d18:1/26:1) | |||
| 7 | 1158.83 | [M + 2H]+2 | 2315.25 (+0.39) | C107H190N4O49 | Acidic (3 isomers) | -Cer(d18:1/26:0) |
| 778.58 | [M + 2H + NH4]+3 | 2315.25 (+0.43) | C107H190N4O49 | Acidic (3 isomers) | -Cer(d18:1/26:0) | |
| [M + 2H + NH4]+3 | 2298.23 (+0.41) | C106H187N5O48 | Lacto/Neolacto (2 isomers) | -Cer(d18:1/26:1) | ||
| 8 | 836.62 | [M + 3H]+3 | 2507.31 (+0.47) | C114H202N4O55 | Neolacto | -Cer(d18:1/24:0) |
| 9 | 955.72 | [M + 2H]+2 | 1909.03 (+0.39) | C88H156N4O40 | Acidic (3 isomers) | -Cer(d18:1/22:0) |
| 10 | 926.70 | [M + 2H]+2 | 1851.05 (+0.33) | C88H158N2O38 | Globo | -Cer(d18:1/26:1) |
| 1851.02 (+0.36) | C86H154N4O38 | Lacto/Neolacto | -Cer(d18:1/20:0) | |||
| 11 | 1129.84 | [M + 2H]+2 | 2257.21 (+0.45) | C104H184N4O48 | Neolacto (2 isomers)/Acidic | -Cer(d18:1/26:1)/-Cer(d18:1/26:0) |
| 759.23 | [M + 2H + NH4]+3 | 2257.21 (+0.45) | -Cer(d18:1/26:1)/-Cer(d18:1/26:0) | |||
| 12 | 897.66 | [M + 2H]+2 | 1793.04 (+0.26) | C86H156N2O36 | Lacto/Neolacto (2 isomers) | -Cer(d18:1/24:0) |
| 13 | 946.722 | [M + H]+ | 945.7116 (+0.0025) | C52H99NO13 | Neutral | -LacCer(d18:1/22:0) |
| 14 | 1100.81 | [M + 2H]+2 | 2199.20 (+0.40) | C102H182N4O46 | Neolacto | -Cer(d18:1/24:0) |
| 739.89 | [M + 2H + NH4]+3 | 2199.20 (+0.41) | -Cer(d18:1/24:0) | |||
| [M + 3H]+3 | 2216.16 (+0.47) | C100H177N5O48 | Lacto/Neolacto (2 isomers) | -Cer(d18:1/20:0) | ||
| 15 | 817.28 | [M − H2O + 3H]+3 | 2467.25 (−0.42) | C110H194N4O56 | Neolacto (2 isomers) | -Cer(d18:1/20:0) |
| [M + 2H + NH4]+3 | 2432.26 (−0.47) | C110H193N5O53 | Neolacto (3 isomers) | -Cer(d18:1/24:1) | ||
| 16 | 1071.79 | [M + 2H]+2 | 2141.16 (+0.40) | C99H176N4O45 | Acidic (9 isomers) | -Cer(d18:1/24:0) |
| 720.54 | [M + 2H + NH4]+3 | 2141.16 (+0.40) | -Cer(d18:1/24:0) | |||
| [M + 3H]+3 | 2158.12 (+0.47) | C97H171N5O47 | Acidic | -Cer(d18:1/20:0) | ||
| 17 | 1042.77 | [M + 2H]+2 | 2083.12 (+0.40) | C96H170N4O44 | Acidic (4 isomers) | -Cer(d18:1/24:0) |
| 701.20 | [M + 2H + NH4]+3 | 2083.12 (+0.42) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castro-Gómez, P.; Montero, O.; Fontecha, J. In-Depth Lipidomic Analysis of Molecular Species of Triacylglycerides, Diacylglycerides, Glycerophospholipids, and Sphingolipids of Buttermilk by GC-MS/FID, HPLC-ELSD, and UPLC-QToF-MS. Int. J. Mol. Sci. 2017, 18, 605. https://doi.org/10.3390/ijms18030605
Castro-Gómez P, Montero O, Fontecha J. In-Depth Lipidomic Analysis of Molecular Species of Triacylglycerides, Diacylglycerides, Glycerophospholipids, and Sphingolipids of Buttermilk by GC-MS/FID, HPLC-ELSD, and UPLC-QToF-MS. International Journal of Molecular Sciences. 2017; 18(3):605. https://doi.org/10.3390/ijms18030605
Chicago/Turabian StyleCastro-Gómez, Pilar, Olimpio Montero, and Javier Fontecha. 2017. "In-Depth Lipidomic Analysis of Molecular Species of Triacylglycerides, Diacylglycerides, Glycerophospholipids, and Sphingolipids of Buttermilk by GC-MS/FID, HPLC-ELSD, and UPLC-QToF-MS" International Journal of Molecular Sciences 18, no. 3: 605. https://doi.org/10.3390/ijms18030605
APA StyleCastro-Gómez, P., Montero, O., & Fontecha, J. (2017). In-Depth Lipidomic Analysis of Molecular Species of Triacylglycerides, Diacylglycerides, Glycerophospholipids, and Sphingolipids of Buttermilk by GC-MS/FID, HPLC-ELSD, and UPLC-QToF-MS. International Journal of Molecular Sciences, 18(3), 605. https://doi.org/10.3390/ijms18030605