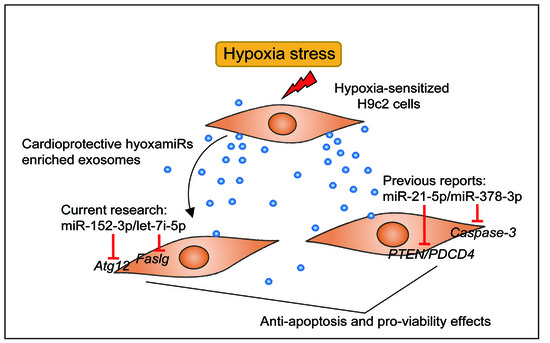
Overexpression of Exosomal Cardioprotective miRNAs Mitigates Hypoxia-Induced H9c2 Cells Apoptosis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Hypoxia Decreases Cell Viability and Induces Apoptosis in H9c2 Cells
2.2. Hypoxia Significantly Modulates hypoxamiR Levels in H9c2 Cells
2.3. Hypoxia Dramatically Alters the miRNAome of H9c2 Cells-Derived Exosomes
2.4. Overexpression of Hypoxia-Induced Exosomal miRNAs Mitigates Hypoxia-Induced Apoptosis
2.5. Atg12 and Faslg Are Targets of miR-152-3p and Let-7i-5p, Respectively
3. Materials and Methods
3.1. Cell Culture and Hypoxia Treatment
3.2. Exosome Isolaton
3.3. Nanoparticle Tracking Analysis
3.4. Small RNA Library Construction and Sequencing
3.5. Prediction and Functional Annotation of Exosomal DE miRNA Targets
3.6. miRNA Transfection
3.7. Detection of Viability and Cell Damage by CCK8 and LDH Release
3.8. Flow Cytometry Analysis of Cell Apoptosis
3.9. Fluorescence Staining of H9c2 Cells with Hoechst 33342/PI
3.10. Dual-Luciferase Activity Assay
3.11. qRT-PCR
3.12. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| DE miRNA | Differentially-Expressed miRNA |
| AMI | Acute Myocardial Infarction |
| HIF | Hypoxia-Inducible Factor |
| HRE | Hypoxia Response Elements |
| CPCs | Cardiac Progenitor Cells |
| CCK8 | Cell Counting Kit-8 |
| LDH | Lactate Dehydrogenase |
| NTA | Nanoparticle Tracking Analysis |
| FBS | Foetal Bovine Serum |
| PBS | Phosphate-Buffered Saline |
| AV | Annexin V-FITC |
| PI | Propidium Iodide |
References
- Members, W.G.; Mozaffarian, D.; Benjamin, E.J.; Go, A.S.; Arnett, D.K.; Blaha, M.J.; Cushman, M.; Das, S.R.; De, F.S.; Després, J.P. Heart Disease and Stroke Statistics-2016 Update: A Report From the American Heart Association. Circulation 2015, 133, e38-360. [Google Scholar]
- Santos-Gallego, C.G.; Picatoste, B.; Badimón, J.J. Pathophysiology of acute coronary syndrome. Curr. Atheroscler. Rep. 2014, 16, 401. [Google Scholar] [CrossRef] [PubMed]
- Heusch, G. Molecular Basis of Cardioprotection Signal Transduction in Ischemic Pre-, Post-, and Remote Conditioning. Circ. Res. 2015, 116, 674–699. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, M.D.; Weil, M.; Raff, M.C.; Jacobson, M.D.; Weil, M.; Raff, M.C. Programmed Cell Death in Animal Development. Cell 1997, 88, 347–354. [Google Scholar] [CrossRef]
- Jiang, B.H.; Rue, E.; Wang, G.L.; Roe, R.; Semenza, G.L. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J. Biol. Chem. 1996, 271, 17771–17778. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Hypoxia-inducible factors in physiology and medicine. Cell 2012, 148, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Nallamshetty, S.; Chan, S.Y.; Loscalzo, J. Hypoxia: A master regulator of microRNA biogenesis and activity. Free Radic. Biol. Med. 2013, 64, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Greijer, A.E.; van de Wall, E. The role of hypoxia inducible factor 1 (HIF-1) in hypoxia induced apoptosis. J. Clin. Pathol. 2004, 57, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Azzouzi, H.E.; Leptidis, S.; Doevendans, P.A.; De Windt, L.J. HypoxamiRs: Regulators of cardiac hypoxia and energy metabolism. Trends Endocrinol. Metab. 2015, 26, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Bang, C.; Batkai, S.; Dangwal, S.; Gupta, S.K.; Foinquinos, A.; Holzmann, A.; Just, A.; Remke, J.; Zimmer, K.; Zeug, A. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J. Clin. Investig. 2014, 124, 2136–2146. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, L.; Li, Y.; Chen, L.; Wang, X.; Guo, W.; Zhang, X.; Qin, G.; He, S.H.; Zimmerman, A. Exosomes/microvesicles from induced pluripotent stem cells deliver cardioprotective miRNAs and prevent cardiomyocyte apoptosis in the ischemic myocardium. Int. J. Cardiol. 2015, 192, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Barile, L.; Lionetti, V.; Cervio, E.; Matteucci, M.; Gherghiceanu, M.; Popescu, L.M.; Torre, T.; Siclari, F.; Moccetti, T.; Vassalli, G. Extracellular vesicles from human cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function after myocardial infarction. Cardiovasc. Res. 2014, 103, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Saraste, A.; Pulkki, K.; Kallajoki, M.; Henriksen, K.; Parvinen, M.; Voipiopulkki, L.M. Apoptosis in human acute myocardial infarction. Circulation 1997, 95, 320–323. [Google Scholar] [CrossRef] [PubMed]
- Bonavita, F.; Stefanelli, C.; Giordano, E.; Columbaro, M.; Facchini, A.; Bonafè, F.; Caldarera, C.M.; Guarnieri, C. H9c2 cardiac myoblasts undergo apoptosis in a model of ischemia consisting of serum deprivation and hypoxia: Inhibition by PMA. FEBS Lett. 2003, 536, 85–91. [Google Scholar] [CrossRef]
- Ekhterae, D.; Lin, Z.; Lundberg, M.S.; Crow, M.T.; Núñez, G. ARC inhibits cytochrome c release from mitochondria and protects against hypoxia-induced apoptosis in heart-derived H9c2 cells. Circ. Res. 1999, 85, 70–77. [Google Scholar] [CrossRef]
- Hu, S.; Huang, M.; Li, Z.; Jia, F.; Ghosh, Z.; Lijkwan, M.A.; Fasanaro, P.; Sun, N.; Wang, X.; Martelli, F.; et al. MicroRNA-210 as a Novel Therapy for Treatment of Ischemic Heart Disease. Circulation 2010, 122, S124–S131. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Qian, L.; Jiang, L. miRNA-dependent cross-talk between VEGF and Ang-2 in hypoxia-induced microvascular dysfunction. Biochem. Biophys. Res. Commun. 2014, 452, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Rane, S.; He, M.; Sayed, D.; Vashistha, H.; Malhotra, A.; Sadoshima, J.; Vatner, D.E.; Vatner, S.F.; Abdellatif, M. Downregulation of miR-199a derepresses hypoxia-inducible factor-1α and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ. Res. 2009, 104, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.J.; Maghsoudi, T.; Wang, T. Exosomes Mediate the Intercellular Communication after Myocardial Infarction. Int. J. Med. Sci. 2016, 13, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Sluijter, J.P.; Verhage, V.; Deddens, J.C.; van den Akker, F.; Doevendans, P.A. Microvesicles and exosomes for intracardiac communication. Cardiovasc. Res. 2014, 102, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Beninson, L.A.; Fleshner, M. Exosomes: An emerging factor in stress-induced immunomodulation. Semin. Immunol. 2014, 26, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Sano, S.; Izumi, Y.; Yamaguchi, T.; Yamazaki, T.; Tanaka, M.; Shiota, M.; Osada-Oka, M.; Nakamura, Y.; Wei, M.; Wanibuchi, H.; et al. Lipid synthesis is promoted by hypoxic adipocyte-derived exosomes in 3T3-L1 cells. Biochem. Biophys. Res. Commun. 2014, 445, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Garcia, N.A.; Ontoria-Oviedo, I.; González-King, H.; Diez-Juan, A.; Sepúlveda, P. Glucose Starvation in Cardiomyocytes Enhances Exosome Secretion and Promotes Angiogenesis in Endothelial Cells. PLoS ONE 2015, 10, e0138849. [Google Scholar] [CrossRef] [PubMed]
- Gray, W.D.; French, K.M.; Ghoshchoudhary, S.; Maxwell, J.T.; Brown, M.E.; Platt, M.O.; Searles, C.D.; Davis, M.E. Identification of therapeutic covariant microRNA clusters in hypoxia-treated cardiac progenitor cell exosomes using systems biology. Circ. Res. 2015, 116, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Haudek, S.B.; Taffet, G.E.; Schneider, M.D.; Mann, D.L. TNF provokes cardiomyocyte apoptosis and cardiac remodeling through activation of multiple cell death pathways. J. Clin. Investig. 2007, 117, 2692–2701. [Google Scholar] [CrossRef] [PubMed]
- Krown, K.A.; Page, M.T.; Nguyen, C.; Zechner, D.; Gutierrez, V.; Comstock, K.L.; Glembotski, C.C.; Quintana, P.J.; Sabbadini, R.A. Tumor necrosis factor alpha-induced apoptosis in cardiac myocytes. Involvement of the sphingolipid signaling cascade in cardiac cell death. J. Clin. Investig. 2011, 98, 2854–2865. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Karin, M. Mammalian MAP kinase signaling cascades. Nature 2001, 410, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Tobiume, K.; Matsuzawa, A.; Takahashi, T.; Nishitoh, H.; Morita, K.I.; Takeda, K.; Minowa, O.; Miyazono, K.; Noda, T.; Ichijo, H. ASK1 is required for sustained activations of JNK/p38 MAP kinases and apoptosis. EMBO Rep. 2001, 2, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.L.; Kumar, S.; Gao, F.; Louden, C.S.; Lopez, B.L.; Christopher, T.A.; Wang, C.; Lee, J.C.; Feuerstein, G.Z.; Yue, T.L. Inhibition of p38 mitogen-activated protein kinase decreases cardiomyocyte apoptosis and improves cardiac function after myocardial ischemia and reperfusion. Circulation 1999, 99, 1685–1691. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Xu, S. ERK1/2 MAP kinases in cell survival and apoptosis. IUBMB Life 2006, 58, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Santos-Gallego, C.G.; Vahl, T.P.; Goliasch, G.; Picatoste, B.; Arias, T.; Ishikawa, K.; Njerve, I.U.; Sanz, J.; Narula, J.; Sengupta, P.P.; et al. Sphingosine-1-Phosphate Receptor Agonist Fingolimod Increases Myocardial Salvage and Decreases Adverse Postinfarction Left Ventricular Remodeling in a Porcine Model of Ischemia/Reperfusion. Circulation 2016, 133, 954–966. [Google Scholar] [CrossRef] [PubMed]
- Castedo, M.; Ferri, K.F.; Kroemer, G. Mammalian target of rapamycin (mTOR): Pro- and anti-apoptotic. Cell Death Differ. 2002, 9, 99–100. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Song, X.W.; Tian, J.; Chen, H.Y.; Li, D.F.; Wang, J.F.; Ren, A.J.; Yuan, W.J.; Lin, L. Overexpression of microRNA-378 attenuates ischemia-induced apoptosis by inhibiting caspase-3 expression in cardiac myocytes. Apoptosis 2012, 17, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.H.; Zhu, P.; Yang, P.; Liu, X.J.; Dong, S.M.; Wang, X.B.; Chun, B.; Zhuang, J.; Zhang, C.X. Ischaemic preconditioning-regulated miR-21 protects heart against ischaemia/reperfusion injury via anti-apoptosis through its target PDCD4. Cardiovasc. Res. 2010, 87, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Buscaglia, L.E.; Li, Y. Apoptosis and the target genes of microRNA-21. Chin. J. Cancer 2011, 30, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Mazière, P.; Enright, A.J. Prediction of microRNA targets. Drug Discov. Today 2007, 12, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: MicroRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, 451–454. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, A.D.; Eisenstein, M.; Ber, Y.; Bialik, S.; Kimchi, A. The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol. Cell 2011, 44, 698–709. [Google Scholar] [CrossRef] [PubMed]
- Setsuta, K.; Seino, Y.; Ogawa, T.; Ohtsuka, T.; Seimiya, K.; Takano, T. Ongoing myocardial damage in chronic heart failure is related to activated tumor necrosis factor and Fas/Fas ligand system. Circ. J. 2004, 68, 747–750. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.; Sata, M.; Lefer, D.J.; Factor, S.M.; Walsh, K.; Kitsis, R.N. Fas pathway is a critical mediator of cardiac myocyte death and MI during ischemia-reperfusion in vivo. Am. J. Physiol. Heart C 2003, 284, H456–H463. [Google Scholar] [CrossRef] [PubMed]
- Barile, L.; Moccetti, T.; Marbán, E.; Vassalli, G. Roles of exosomes in cardioprotection. Eur. Heart J. 2016. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, S.; Losordo, D.W. Exosomes and Cardiac Repair After Myocardial Infarction. Circ. Res. 2014, 114, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Vicencio, J.M.; Yellon, D.M.; Sivaraman, V.; Das, D.; Boi-Doku, C.; Arjun, S.; Zheng, Y.; Riquelme, J.A.; Kearney, J.; Sharma, V. Plasma exosomes protect the myocardium from ischemia-reperfusion injury. J. Am. Coll. Cardiol. 2015, 65, 1525–1536. [Google Scholar] [CrossRef] [PubMed]
- Skyschally, A.; Gent, S.; Amanakis, G.; Schulte, C.; Kleinbongard, P.; Heusch, G. Across-Species Transfer of Protection by Remote Ischemic Preconditioning With Species-Specific Myocardial Signal Transduction by Reperfusion Injury Salvage Kinase and Survival Activating Factor Enhancement Pathways Novelty and Significance. Circ. Res. 2015, 117, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Osanai, T.; Nishizaki, F.; Sukekawa, T. Matrix metalloprotein-9 activation under cell-to-cell interaction between endothelial cells and monocytes: Possible role of hypoxia and tumor necrosis factor-α. Heart Vessels 2012, 27, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.R.; Martinsmarques, T.; Ribeirorodrigues, T.; Ferreira, J.V.; Catarino, S.; Pinho, M.J.; Zuzarte, M.; Anjo, S.I.; Manadas, B.; Sluijter, J.P.G.; et al. Gap junctional protein Cx43 is involved in the communication between extracellular vesicles and mammalian cells. Sci. Rep. 2015, 5, 13243. [Google Scholar] [CrossRef] [PubMed]
- Soo, C.Y.; Song, Y.; Zheng, Y.; Campbell, E.C.; Riches, A.C.; Gunnmoore, F.; Powis, S.J. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology 2012, 136, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Livshts, M.A.; Khomyakova, E.; Evtushenko, E.G.; Lazarev, V.N.; Kulemin, N.A.; Semina, S.E.; Generozov, E.V.; Govorun, V.M. Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci. Rep. 2015, 6, 21447. [Google Scholar] [CrossRef] [PubMed]





© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Ma, J.; Long, K.; Qiu, W.; Wang, Y.; Hu, Z.; Liu, C.; Luo, Y.; Jiang, A.; Jin, L.; et al. Overexpression of Exosomal Cardioprotective miRNAs Mitigates Hypoxia-Induced H9c2 Cells Apoptosis. Int. J. Mol. Sci. 2017, 18, 711. https://doi.org/10.3390/ijms18040711
Zhang J, Ma J, Long K, Qiu W, Wang Y, Hu Z, Liu C, Luo Y, Jiang A, Jin L, et al. Overexpression of Exosomal Cardioprotective miRNAs Mitigates Hypoxia-Induced H9c2 Cells Apoptosis. International Journal of Molecular Sciences. 2017; 18(4):711. https://doi.org/10.3390/ijms18040711
Chicago/Turabian StyleZhang, Jinwei, Jideng Ma, Keren Long, Wanling Qiu, Yujie Wang, Zihui Hu, Can Liu, Yi Luo, Anan Jiang, Long Jin, and et al. 2017. "Overexpression of Exosomal Cardioprotective miRNAs Mitigates Hypoxia-Induced H9c2 Cells Apoptosis" International Journal of Molecular Sciences 18, no. 4: 711. https://doi.org/10.3390/ijms18040711
APA StyleZhang, J., Ma, J., Long, K., Qiu, W., Wang, Y., Hu, Z., Liu, C., Luo, Y., Jiang, A., Jin, L., Tang, Q., Wang, X., Li, X., & Li, M. (2017). Overexpression of Exosomal Cardioprotective miRNAs Mitigates Hypoxia-Induced H9c2 Cells Apoptosis. International Journal of Molecular Sciences, 18(4), 711. https://doi.org/10.3390/ijms18040711