Structure, Expression, and Functional Analysis of the Hexokinase Gene Family in Cassava
Abstract
:1. Introduction
2. Results
2.1. Identification of MeHXK Family Members
2.2. Structural Analysis of MeHXK1–7
2.3. Evolutionary Analysis of MeHXKs
2.4. Motif Distribution in MeHXKs
2.5. 3D Structures of MeHXKs
2.6. Differential Expression Analysis of MeHXK1–7 in Cassava Organs or Tissues
2.7. Differential Expression of MeHXK Genes during Cassava Tuber Root Development
2.8. The Activity of HXKs in Cassava Tubers during Tuber Root Development
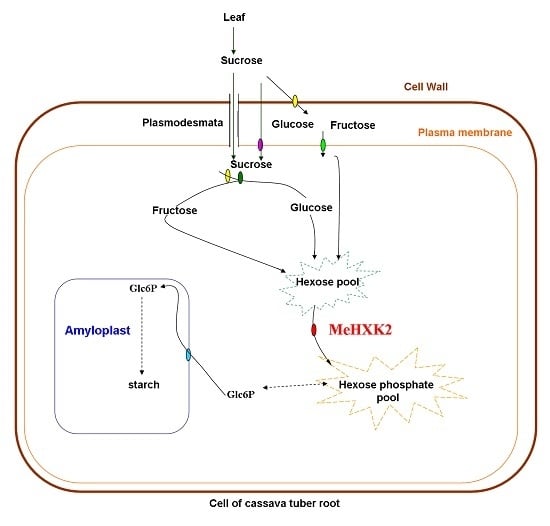
2.9. Yeast Complementation of MeHXK2
2.10. Kinetic Analysis of MeHXK2
3. Discussion
3.1. Identification and Characterization of MeHXK Genes
3.2. Differential Expression and Enzymatic Activity of MeHXK1–7
4. Materials and Methods
4.1. Plant Materials
4.2. Cloning of Full-Length MeHXKs
4.3. Sequence Analysis of MeHXKs
4.4. Homology Modeling of MeHXKs
4.5. qPCR Analysis
4.6. Activity Analysis of HXKs
4.7. Yeast Complementation and Enzymatic Analysis of MeHXK2
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Granot, D.; Kelly, G.; Stein, O.; David-Schwartz, R. Substantial roles of hexokinase and fructokinase in the effects of sugars on plant physiology and development. J. Exp. Bot. 2014, 65, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Siemens, J.; GonzáLez, M.C.; Wolf, S.; Hofmann, C.; Greiner, S.; Du, Y.; Rausch, T.; Roitsch, T.; Ludwig-MüLler, J. Extracellular invertase is involved in the regulation of clubroot disease in Arabidopsis thaliana. Mol. Plant Pathol. 2011, 12, 247–262. [Google Scholar] [CrossRef] [PubMed]
- David-Schwartz, R.; Weintraub, L.; Vidavski, R.; Zemach, H.; Murakhovsky, L.; Swartzberg, D.; Granot, D. The SLFRK4 promoter is active only during late stages of pollen and anther development. Plant Sci. 2013, 199, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Granot, D. Role of tomato hexose kinases. Funct. Plant Biol. 2007, 34, 564–570. [Google Scholar] [CrossRef]
- Granot, D. Putting plant hexokinases in their proper place. Phytochemistry 2008, 69, 2649–2654. [Google Scholar] [CrossRef] [PubMed]
- Karve, A.; Rauh, B.L.; Xia, X.; Kandasamy, M.; Meagher, R.B.; Sheen, J.; Moore, B.D. Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta 2008, 228, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Sheen, J. Master regulators in plant glucose signaling networks. J. Plant Biol. 2014, 57, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Olsson, T.; Thelander, M.; Ronne, H. A novel type of chloroplast stromal hexokinase is the major glucose-phosphorylating enzyme in the moss physcomitrella patens. J. Biol. Chem. 2003, 278, 44439–44447. [Google Scholar] [CrossRef] [PubMed]
- Granot, D.; David-Schwartz, R.; Kelly, G. Hexose kinases and their role in sugar-sensing and plant development. Front. Plant Sci. 2013, 4, 44. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Zhang, H.; Zhou, X.; Liu, H.; Liu, Y.; Li, J.; Han, S.; Wang, Y. Subcellular localization of rice hexokinase (OsHXK) family members in the mesophyll protoplasts of tobacco. Biol. Plant. 2011, 55, 173–177. [Google Scholar] [CrossRef]
- Xu, F.Q.; Li, X.R.; Ruan, Y.L. RNAi-mediated suppression of hexokinase gene OSHXK10 in rice leads to non-dehiscent anther and reduction of pollen germination. Plant Sci. 2008, 175, 674–684. [Google Scholar] [CrossRef]
- Lugassi, N.; Gilor Kelly, L.F.; Yaniv, Y.; Attia, Z.; Levi, A.; Alchanatis, V.; Moshelion, M.; Raveh, E.; Carmi, N.; Granot, D. Expression of Arabidopsis hexokinase in citrus guard cells controls stomatal aperture and reduces transpiration. Front. Plant Sci. 2015, 6, 1114. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.B.; Cho, J.I.; Ryoo, N.; Shin, D.H.; Park, Y.I.; Hwang, Y.S.; Lee, S.K.; An, G.; Jeon, J.S. Role of rice cytosolic hexokinase OSHXK7 in sugar signaling and metabolism. J. Integr. Plant Biol. 2015, 58, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Godbole, A.; Dubey, A.K.; Reddy, P.S.; Udayakumar, M.; Mathew, M.K. Mitochondrial VDAC and hexokinase together modulate plant programmed cell death. Protoplasma 2013, 250, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Sarowar, S.; Lee, J.Y.; Ahn, E.R.; Pai, H.S. A role of hexokinases in plant resistance to oxidative stress and pathogen infection. J. Plant Biol. 2008, 51, 341–346. [Google Scholar] [CrossRef]
- Singh, M.; Gupta, A.; Laxmi, A. Glucose control of root growth direction in Arabidopsis thaliana. J. Plant Biol. 2014, 65, 2981–2993. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.M.; Heinzel, N.; Giese, J.O.; Koeber, J.; Melzer, M.; Rutten, T.; von Wiren, N.; Sonnewald, U.; Hajirezaei, M.R. A dual role of tobacco hexokinase 1 in primary metabolism and sugar sensing. Plant Cell Environ. 2013, 36, 1311–1327. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.I.; Ryoo, N.; Ko, S.; Lee, S.K.; Lee, J.; Jung, K.H.; Lee, Y.H.; Bhoo, S.H.; Winderickx, J.; An, G. Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.). Planta 2006, 224, 598–611. [Google Scholar] [CrossRef] [PubMed]
- Jansson, C.; Westerbergh, A.; Zhang, J.; Hu, X.; Sun, C. Cassava, a potential biofuel crop in (the) people’s republic of china. Appl. Energy 2009, 86, S95–S99. [Google Scholar] [CrossRef]
- Polit, J.T.; Ciereszko, I. In situ activities of hexokinase and fructokinase in relation to phosphorylation status of root meristem cells of Vicia faba during reactivation from sugar starvation. Physiol. Plant. 2009, 135, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Sheen, J.; Jang, J.C. The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol. Biol. 2000, 44, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Q.; Li, L.M.; Yang, P.P.; Gong, C.L. The role of hexokinases from grape berries (Vitis vinifera L.) in regulating the expression of cell wall invertase and sucrose synthase genes. Plant Cell Rep. 2014, 33, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Dian, W.; Jiang, H.; Chen, Q.; Liu, F.; Wu, P. Cloning and characterization of the granule-bound starch synthase II gene in rice: Gene expression is regulated by the nitrogen level, sugar and circadian rhythm. Planta 2003, 218, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Axel, T.; Katrin, P.; Anja, B.; Natalia, P.; Rowan, M.K.; Halford, N.G.; Peter, G. Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signalling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J. 2003, 35, 490–500. [Google Scholar]
- Maruthi, M.N.; Bouvaine, S.; Tufan, H.A.; Mohammed, I.U.; Hillocks, R.J. Transcriptional response of virus-infected cassava and identification of putative sources of resistance for cassava brown streak disease. PLoS ONE 2014, 9, e96642. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, N.N.; Ihemere, U.; Ellery, C.; Sayre, R.T. Overexpression of hydroxynitrile lyase in cassava roots elevates protein and free amino acids while reducing residual cyanogen levels. PLoS ONE 2011, 6, e21996. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Z.; Zhao, J.Y.; Wu, S.M.; Fan, X.W.; Luo, X.L.; Chen, B.S. Characters related to higher starch accumulation in cassava storage roots. Sci. Rep. 2016, 6, 19823. [Google Scholar] [CrossRef] [PubMed]
- Baguma, Y.; Sun, C.; Borén, M.; Olsson, H.; Rosenqvist, S.; Mutisya, J.; Rubaihayo, P.R.; Jansson, C. Sugar-mediated semidian oscillation of gene expression in the cassava storage root regulates starch synthesis. Plant Signal. Behav. 2008, 3, 439. [Google Scholar] [CrossRef] [PubMed]
- Troncoso-Ponce, M.; Rivoal, J.; Dorion, S.; Moisan, M.-C.; Garcés, R.; Martínez-Force, E. Cloning, biochemical characterization and expression of a sunflower (Helianthus annuus L.) hexokinase associated with seed storage compounds accumulation. J. Plant Physiol. 2011, 168, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Kuser, P.R.; Krauchenco, S.; Antunes, O.A.; Polikarpov, I. The high resolution crystal structure of yeast hexokinase PII with the correct primary sequence provides new insights into its mechanism of action. J. Biol. Chem. 2000, 275, 20814–20821. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 7, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, J.; Chen, Y.; Li, R.; Wang, H.; Ding, L.; Wei, J. Isolation, structural analysis, and expression characteristics of the maize (Zea mays L.) hexokinase gene family. Mol. Biol. Rep. 2014, 41, 6157–6166. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, A.; Olsson, T.; Ulfstedt, M.; Thelander, M.; Ronne, H. Two novel types of hexokinases in the moss Physcomitrella patens. BMC Plant Biol. 2011, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Kandel-Kfir, M.; Damari-Weissler, H.; German, M.; Gidoni, D.; Mett, A.; Belausov, E.; Petreikov, M.; Adir, N.; Granot, D. Two newly identified membrane-associated and plastidic tomato HXKs: Characteristics, predicted structure and intracellular localization. Planta 2006, 224, 1341–1352. [Google Scholar] [CrossRef] [PubMed]
- Karve, A.; Moore, B.D. Function of Arabidopsis hexokinase-like1 as a negative regulator of plant growth. J. Exp. Bot. 2009, 60, 4137–4149. [Google Scholar] [CrossRef] [PubMed]
- Dai, N.; Kandel-Kfir, M.; Petreikov, M.; Hanael, R.; Levin, I.; Ricard, B.; Rothan, C.; Schaffer, A.A.; Granot, D. The tomato hexokinase LeHXK1 cloning, mapping, expression pattern and phylogenetic relationships. Plant Sci. 2002, 163, 581–590. [Google Scholar] [CrossRef]
- Prochnik, S.; Marri, P.R.; Desany, B.; Rabinowicz, P.D.; Kodira, C.; Mohiuddin, M.; Rodriguez, F.; Fauquet, C.; Tohme, J.; Harkins, T. The cassava genome: Current progress, future directions. Trop. Plant Biol. 2012, 5, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Richmond, T. Topology prediction of membrane proteins. Genome Biol. 2000, 1, 224. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]













| Gene Name | Accession Number | Open Reading Frame (ORF) Lengths (bp) | Protein Lengths (a.a) | pI | Mw (Da) |
|---|---|---|---|---|---|
| MeHXK1 | KJ417433 | 1488 | 496 | 6.20 | 53,356.2 |
| MeHXK2 | KJ417434 | 1494 | 498 | 6.01 | 53,901.4 |
| MeHXK3 | KJ417435 | 1524 | 508 | 6.91 | 55,303.8 |
| MeHXK4 | KJ417436 | 1491 | 497 | 7.70 | 54,364.9 |
| MeHXK5 | KJ417437 | 1482 | 494 | 5.81 | 53,440.8 |
| MeHXK6 | KJ417438 | 861 | 287 | 7.05 | 30,714.7 |
| MeHXK7 | KJ417439 | 933 | 311 | 6.43 | 33,671.1 |
| Substrate | Km (mM) | Vmax (nmol/mg·pr/min) | Km/Vmax |
|---|---|---|---|
| Glucose | 0.078 | 228.1 | 33.59 |
| Fructose | 1.321 | 644.0 | 2.05 |
| Gene | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) |
|---|---|---|
| MeHXK1 | TTTCACGTTCTTTGATCTCACCG | TTAGCCAATGAATCCCAGTTGC |
| MeHXK2 | GCAGCTAGTGGTCATGGGAAAGG | TTTCACATACCCAATTTCGAGTTAGC |
| MeHXK3 | GAAACGGCGACGTTATGGTTGA | ATTTCATCTTTCCAGGTGGGTT |
| MeHXK4 | GTAGTGCTGATGGGAAGGGTGA | TGGATAGCGTACAGTAGCTATAAA |
| MeHXK5 | TATCACAATCAACCTTCTTTCTTCA | ATTCGCATTATCGTTAAGTTGAGG |
| MeHXK6 | GTACTCATGGGAAAGGTGGCG | ATAAGCCGAATGAAAAAGGATGAC |
| MeHXK7 | CTCTTGTTGTAGTGAAGGTTTTGA | ATCAGTTTACTCATTAGGAATCAGG |
| Gene | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) |
|---|---|---|
| MeHXK1 | CATTGGACGGAGGGCTGTTT | GTCATCCTTTTTCATTCGGCTTAT |
| MeHXK2 | ATGGTCTTCTTCCTAAATCTGGG | GTGGAACAACATCACCAAAGAAG |
| MeHXK3 | GCGACGTTATGGTTGAGGTTAT | ACTCTTCCAACTCCCTCAACAC |
| MeHXK4 | GCGGTGGCGATGAGTGTC | CTCCAACTCCCTCAATAATCCC |
| MeHXK5 | AGTATGTGCTACCGCTGTGGC | CCCACTTATCACCCGTCCTCA |
| MeHXK6 | TCTAACATCGTCTCGGTGGCT | ACAATCCTTTCTCGTTCCCGC |
| MeHXK7 | TTACTTCACTGCCCAGCGGACTT | TCTTGCGGTTGCTTCACCATCAT |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geng, M.-T.; Yao, Y.; Wang, Y.-L.; Wu, X.-H.; Sun, C.; Li, R.-M.; Fu, S.-P.; Duan, R.-J.; Liu, J.; Hu, X.-W.; et al. Structure, Expression, and Functional Analysis of the Hexokinase Gene Family in Cassava. Int. J. Mol. Sci. 2017, 18, 1041. https://doi.org/10.3390/ijms18051041
Geng M-T, Yao Y, Wang Y-L, Wu X-H, Sun C, Li R-M, Fu S-P, Duan R-J, Liu J, Hu X-W, et al. Structure, Expression, and Functional Analysis of the Hexokinase Gene Family in Cassava. International Journal of Molecular Sciences. 2017; 18(5):1041. https://doi.org/10.3390/ijms18051041
Chicago/Turabian StyleGeng, Meng-Ting, Yuan Yao, Yun-Lin Wang, Xiao-Hui Wu, Chong Sun, Rui-Mei Li, Shao-Ping Fu, Rui-Jun Duan, Jiao Liu, Xin-Wen Hu, and et al. 2017. "Structure, Expression, and Functional Analysis of the Hexokinase Gene Family in Cassava" International Journal of Molecular Sciences 18, no. 5: 1041. https://doi.org/10.3390/ijms18051041
APA StyleGeng, M. -T., Yao, Y., Wang, Y. -L., Wu, X. -H., Sun, C., Li, R. -M., Fu, S. -P., Duan, R. -J., Liu, J., Hu, X. -W., & Guo, J. -C. (2017). Structure, Expression, and Functional Analysis of the Hexokinase Gene Family in Cassava. International Journal of Molecular Sciences, 18(5), 1041. https://doi.org/10.3390/ijms18051041