Sensitivity of HOXB13 as a Diagnostic Immunohistochemical Marker of Prostatic Origin in Prostate Cancer Metastases: Comparison to PSA, Prostein, Androgen Receptor, ERG, NKX3.1, PSAP, and PSMA
Abstract
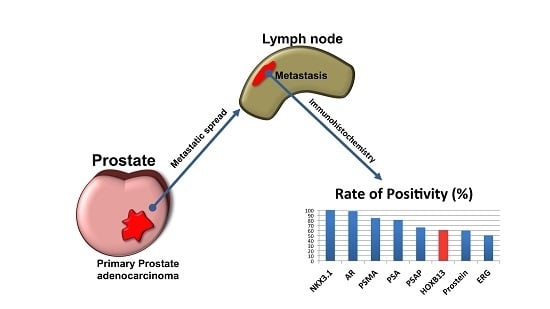
:1. Introduction
2. Results
2.1. Immunohistochemical Staining Patterns
2.2. Statistical Evaluation of Prostate Markers
3. Material and Methods
3.1. Case Selection and Construction of Tissue Microarray
3.2. Immunohistochemistry
3.3. Evaluation of Immunohistochemistry
3.4. Statistics
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ablin, R.J.; Bronson, P.; Soanes, W.A.; Witebsky, E. Tissue- and species-specific antigens of normal human prostatic tissue. J. Immunol. 1970, 104, 1329–1339. [Google Scholar] [PubMed]
- Frankel, A.E.; Rouse, R.V.; Wang, M.C.; Chu, T.M.; Herzenberg, L.A. Monoclonal antibodies to a human prostate antigen. Cancer Res. 1982, 42, 3714–3718. [Google Scholar] [PubMed]
- Steffens, J.; Friedmann, W.; Lobeck, H. Immunohistochemical diagnosis of the metastasizing prostatic carcinoma. Eur. Urol. 1985, 11, 91–94. [Google Scholar] [PubMed]
- Stein, B.S.; Vangore, S.; Petersen, R.O.; Kendall, A.R. Immunoperoxidase localization of prostate-specific antigen. Am. J. Surg. Pathol. 1982, 6, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Epstein, J.I.; Egevad, L.; Humphrey, P.A.; Montironi, R.; Members of the ISUP Immunohistochemistry in Diagnostic Urologic Pathology Group. Best practices recommendations in the application of immunohistochemistry in the prostate: Report from the international society of urologic pathology consensus conference. Am. J. Surg. Pathol. 2014, 38, e6–e19. [Google Scholar] [PubMed]
- Kalos, M.; Askaa, J.; Hylander, B.L.; Repasky, E.A.; Cai, F.; Vedvick, T.; Reed, S.G.; Wright, G.L., Jr.; Fanger, G.R. Prostein expression is highly restricted to normal and malignant prostate tissues. Prostate 2004, 60, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Kalos, M.; Stolk, J.A.; Zasloff, E.J.; Zhang, X.; Houghton, R.L.; Filho, A.M.; Nolasco, M.; Badaro, R.; Reed, S.G. Identification and characterization of prostein, a novel prostate-specific protein. Cancer Res. 2001, 61, 1563–1568. [Google Scholar] [PubMed]
- Yin, M.; Dhir, R.; Parwani, A.V. Diagnostic utility of p501s (prostein) in comparison to prostate specific antigen (PSA) for the detection of metastatic prostatic adenocarcinoma. Diagn. Pathol. 2007, 2, 41. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, T.; Herawi, M.; Epstein, J.I.; Illei, P.B. The role of p501s and PSA in the diagnosis of metastatic adenocarcinoma of the prostate. Am. J. Surg. Pathol. 2007, 31, 1351–1355. [Google Scholar] [CrossRef] [PubMed]
- He, W.W.; Sciavolino, P.J.; Wing, J.; Augustus, M.; Hudson, P.; Meissner, P.S.; Curtis, R.T.; Shell, B.K.; Bostwick, D.G.; Tindall, D.J.; et al. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics 1997, 43, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Bowen, C.; Bubendorf, L.; Voeller, H.J.; Slack, R.; Willi, N.; Sauter, G.; Gasser, T.C.; Koivisto, P.; Lack, E.E.; Kononen, J.; et al. Loss of NKX3.1 expression in human prostate cancers correlates with tumor progression. Cancer Res. 2000, 60, 6111–6115. [Google Scholar] [PubMed]
- Edwards, S.; Campbell, C.; Flohr, P.; Shipley, J.; Giddings, I.; Te-Poele, R.; Dodson, A.; Foster, C.; Clark, J.; Jhavar, S.; et al. Expression analysis onto microarrays of randomly selected cDNA clones highlights HOXB13 as a marker of human prostate cancer. Br. J. Cancer 2005, 92, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Varinot, J.; Cussenot, O.; Roupret, M.; Conort, P.; Bitker, M.O.; Chartier-Kastler, E.; Cheng, L.; Comperat, E. HOXB13 is a sensitive and specific marker of prostate cells, useful in distinguishing between carcinomas of prostatic and urothelial origin. Virchows Arch. 2013, 463, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Zabalza, C.V.; Adam, M.; Burdelski, C.; Wilczak, W.; Wittmer, C.; Kraft, S.; Krech, T.; Steurer, S.; Koop, C.; Hube-Magg, C.; et al. HOXB13 overexpression is an independent predictor of early PSA recurrence in prostate cancer treated by radical prostatectomy. Oncotarget 2015, 6, 12822–12834. [Google Scholar] [CrossRef] [PubMed]
- Barresi, V.; Ieni, A.; Cardia, R.; Licata, L.; Vitarelli, E.; Reggiani Bonetti, L.; Tuccari, G. HOXB13 as an immunohistochemical marker of prostatic origin in metastatic tumors. APMIS 2016, 124, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Varinot, J.; Furudoi, A.; Drouin, S.; Phe, V.; Penna, R.R.; Roupret, M.; Bitker, M.O.; Cussenot, O.; Comperat, E. HOXB13 protein expression in metastatic lesions is a promising marker for prostate origin. Virchows Arch. 2016, 468, 619–622. [Google Scholar] [CrossRef] [PubMed]
- Remmele, W.; Stegner, H.E. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Der. Pathol. 1987, 8, 138–140. [Google Scholar]
- Howarth, D.J.; Aronson, I.B.; Diamandis, E.P. Immunohistochemical localization of prostate-specific antigen in benign and malignant breast tissues. Br. J. Cancer 1997, 75, 1646–1651. [Google Scholar] [CrossRef] [PubMed]
- Ide, H.; Inoue, S.; Miyamoto, H. Histopathological and prognostic significance of the expression of sex hormone receptors in bladder cancer: A meta-analysis of immunohistochemical studies. PLoS ONE 2017, 12, e0174746. [Google Scholar] [CrossRef] [PubMed]
- Udager, A.M.; Chiosea, S.I. Salivary duct carcinoma: An update on morphologic mimics and diagnostic use of androgen receptor immunohistochemistry. Head Neck Pathol. 2017. [Google Scholar] [CrossRef] [PubMed]
- van Leenders, G.J.; Boormans, J.L.; Vissers, C.J.; Hoogland, A.M.; Bressers, A.A.; Furusato, B.; Trapman, J. Antibody EPR3864 is specific for ERG genomic fusions in prostate cancer: Implications for pathological practice. Mod. Pathol. 2011, 24, 1128–1138. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, M.; Wang, Z.F.; Paetau, A.; Tan, S.H.; Dobi, A.; Srivastava, S.; Sesterhenn, I. ERG transcription factor as an immunohistochemical marker for vascular endothelial tumors and prostatic carcinoma. Am. J. Surg. Pathol. 2011, 35, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Kuda, M.; Kohashi, K.; Yamamoto, H.; Takemoto, J.; Ishii, T.; Iura, K.; Maekawa, A.; Bekki, H.; Ito, T.; et al. Histological and immunohistochemical characteristics of undifferentiated small round cell sarcomas associated with CIC-DUX4 and BCOR-CCNB3 fusion genes. Virchows Arch. 2017, 470, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Naughton, D.; Busam, K.; Pulitzer, M. ERG is a useful immunohistochemical marker to distinguish leukemia cutis from nonneoplastic leukocytic infiltrates in the skin. Am. J. Dermatopathol. 2016, 38, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Chuang, A.Y.; DeMarzo, A.M.; Veltri, R.W.; Sharma, R.B.; Bieberich, C.J.; Epstein, J.I. Immunohistochemical differentiation of high-grade prostate carcinoma from urothelial carcinoma. Am. J. Surg. Pathol. 2007, 31, 1246–1255. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, M.; Parwani, A.V. Diagnostic utility of p63/P501S double sequential immunohistochemical staining in differentiating urothelial carcinoma from prostate carcinoma. Diagn. Pathol. 2011, 6, 67. [Google Scholar] [CrossRef] [PubMed]
- Perner, S.; Rupp, N.J.; Braun, M.; Rubin, M.A.; Moch, H.; Dietel, M.; Wernert, N.; Jung, K.; Stephan, C.; Kristiansen, G. Loss of SLC45A3 protein (prostein) expression in prostate cancer is associated with SLC45A3-ERG gene rearrangement and an unfavorable clinical course. Int. J. Cancer 2013, 132, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Bostwick, D.G.; Pacelli, A.; Blute, M.; Roche, P.; Murphy, G.P. Prostate specific membrane antigen expression in prostatic intraepithelial neoplasia and adenocarcinoma: A study of 184 cases. Cancer 1998, 82, 2256–2261. [Google Scholar] [CrossRef]
- Silver, D.A.; Pellicer, I.; Fair, W.R.; Heston, W.D.; Cordon-Cardo, C. Prostate-specific membrane antigen expression in normal and malignant human tissues. Clin. Cancer Res. 1997, 3, 81–85. [Google Scholar] [PubMed]
- Gurel, B.; Ali, T.Z.; Montgomery, E.A.; Begum, S.; Hicks, J.; Goggins, M.; Eberhart, C.G.; Clark, D.P.; Bieberich, C.J.; Epstein, J.I.; et al. NKX3.1 as a marker of prostatic origin in metastatic tumors. Am. J. Surg. Pathol. 2010, 34, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Gelmann, E.P.; Bowen, C.; Bubendorf, L. Expression of NKX3.1 in normal and malignant tissues. Prostate 2003, 55, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Skotheim, R.I.; Korkmaz, K.S.; Klokk, T.I.; Abeler, V.M.; Korkmaz, C.G.; Nesland, J.M.; Fossa, S.D.; Lothe, R.A.; Saatcioglu, F. NKX3.1 expression is lost in testicular germ cell tumors. Am. J. Pathol. 2003, 163, 2149–2154. [Google Scholar] [CrossRef]
- Conner, J.R.; Hornick, J.L. Metastatic carcinoma of unknown primary: Diagnostic approach using immunohistochemistry. Adv. Anat. Pathol. 2015, 22, 149–167. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Ke, J.Q.; Jiang, F.Z.; Wang, X.J.; Wang, F.Y.; Li, Y.R.; Lu, W.; Wan, X.P. Tumor-associated macrophage-derived CXCL8 could induce ERα suppression via HOXB13 in endometrial cancer. Cancer lette. 2016, 376, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.; Sun, Q.K.; Wang, W.; Jia, W.D. High-level expression OF HOXB13 Is closely associated with tumor angiogenesis and poor prognosis of hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 2925–2933. [Google Scholar] [PubMed]
- Zhai, L.L.; Wu, Y.; Cai, C.Y.; Tang, Z.G. Overexpression of homeobox B-13 correlates with angiogenesis, aberrant expression of emt markers, aggressive characteristics and poor prognosis in pancreatic carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 6919–6927. [Google Scholar] [PubMed]
- Queisser, A.; Hagedorn, S.A.; Braun, M.; Vogel, W.; Duensing, S.; Perner, S. Comparison of different prostatic markers in lymph node and distant metastases of prostate cancer. Mod. Pathol. 2015, 28, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Gown, A.M. Diagnostic immunohistochemistry: What can go wrong and how to prevent it. Arch. Pathol. Lab. Med. 2016, 140, 893–898. [Google Scholar] [CrossRef] [PubMed]

| Marker | Detection Rate (%) | Mean IRS | Number of Cases |
|---|---|---|---|
| PSA | 80.8 | 6.3 | 52 |
| PSAP | 66.0 | 3.5 | 53 |
| PSMA | 84.3 | 6.0 | 51 |
| Prostein | 59.6 | 4.2 | 52 |
| Androgen receptor (AR) | 98.1 | 6.7 | 53 |
| ERG | 50.0 | 2.6 | 52 |
| NKX3.1 | 100.0 | 8.0 | 50 |
| HOXB13 | 60.4 | 4.7 | 53 |
| Antigen | Clone | Provider | Dilution | Platform | Protocol |
|---|---|---|---|---|---|
| PSA | Polyclonal, rabbit | DAKO | 1:20,000 | Autostainer | No pretreatment |
| PSAP | PASE/4LJ | Cell Marque | 1:6000 | Autostainer | HIER (pH 6, 20 s 98 ℃) |
| PSMA | 3E6 | DAKO | 1:500 | Benchmark | CC1 (pH 8), ultraview |
| Prostein | 10E3 | DAKO | 1:100 | Benchmark | CC1 (pH 8), ultraview |
| Androgen Receptor (AR) | AR441 | DAKO | 1:400 | Autostainer | HIER (pH 6, 20 s 98 ℃) |
| ERG | EPR3864 | Biologo | 1:100 | Autostainer | HIER (pH 6, 20 s 98 ℃) |
| NKX3.1 | EP356 | Cell Marque | 1:200 | Benchmark | CC1 (pH 8), ultraview |
| HOXB13 | F-9 | Santa Cruz | 1:50 | Benchmark | CC1 (pH 8), optiview |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kristiansen, I.; Stephan, C.; Jung, K.; Dietel, M.; Rieger, A.; Tolkach, Y.; Kristiansen, G. Sensitivity of HOXB13 as a Diagnostic Immunohistochemical Marker of Prostatic Origin in Prostate Cancer Metastases: Comparison to PSA, Prostein, Androgen Receptor, ERG, NKX3.1, PSAP, and PSMA. Int. J. Mol. Sci. 2017, 18, 1151. https://doi.org/10.3390/ijms18061151
Kristiansen I, Stephan C, Jung K, Dietel M, Rieger A, Tolkach Y, Kristiansen G. Sensitivity of HOXB13 as a Diagnostic Immunohistochemical Marker of Prostatic Origin in Prostate Cancer Metastases: Comparison to PSA, Prostein, Androgen Receptor, ERG, NKX3.1, PSAP, and PSMA. International Journal of Molecular Sciences. 2017; 18(6):1151. https://doi.org/10.3390/ijms18061151
Chicago/Turabian StyleKristiansen, Ilka, Carsten Stephan, Klaus Jung, Manfred Dietel, Anja Rieger, Yuri Tolkach, and Glen Kristiansen. 2017. "Sensitivity of HOXB13 as a Diagnostic Immunohistochemical Marker of Prostatic Origin in Prostate Cancer Metastases: Comparison to PSA, Prostein, Androgen Receptor, ERG, NKX3.1, PSAP, and PSMA" International Journal of Molecular Sciences 18, no. 6: 1151. https://doi.org/10.3390/ijms18061151
APA StyleKristiansen, I., Stephan, C., Jung, K., Dietel, M., Rieger, A., Tolkach, Y., & Kristiansen, G. (2017). Sensitivity of HOXB13 as a Diagnostic Immunohistochemical Marker of Prostatic Origin in Prostate Cancer Metastases: Comparison to PSA, Prostein, Androgen Receptor, ERG, NKX3.1, PSAP, and PSMA. International Journal of Molecular Sciences, 18(6), 1151. https://doi.org/10.3390/ijms18061151