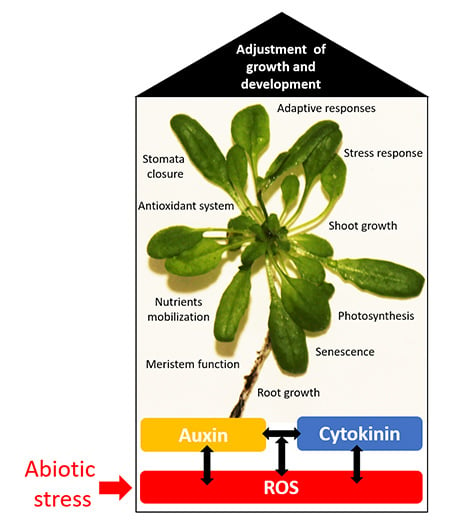
Plants under Stress: Involvement of Auxin and Cytokinin
Abstract
:1. Introduction
2. Role of Auxin and Cytokinin during Plant Response to Abiotic Stress
2.1. Targets of Auxin and Cytokinin Biosynthesis Modulated by Stress
2.2. Stress-Induced Modulation of Auxin and Cytokinin Metabolic Routes
2.3. Modulation of Auxin and Cytokinin Transport by Stress
2.4. Auxin and Cytokinin Signaling Circuits Influenced by Stress
3. Auxin–Cytokinin Crosstalk
3.1. Metabolism-Related Auxin-Cytokinin Crosstalk Components
3.2. Signaling-Related Auxin-Cytokinin Crosstalk Components
4. Abiotic Stress-Auxin-Cytokinin Transcriptional Crosstalk Networks
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Skoog, F.; Miller, C.O. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 1957, 54, 118–130. [Google Scholar]
- Benjamins, R.; Scheres, B. Auxin: The Looping Star in Plant Development. Annu. Rev. Plant Biol. 2008, 59, 443–465. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.W.; Werr, W. Cytokinin–auxin crosstalk in cell type specification. Trends Plant Sci. 2015, 20, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Del Bianco, M.; Giustini, L.; Sabatini, S. Spatiotemporal changes in the role of cytokinin during root development. New Phytol. 2013, 199, 324–338. [Google Scholar] [CrossRef] [PubMed]
- Muller, D.; Leyser, O. Auxin, cytokinin and the control of shoot branching. Ann. Bot. 2011, 107, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Robert, H.S.; Crhak Khaitova, L.; Mroue, S.; Benková, E. The importance of localized auxin production for morphogenesis of reproductive organs and embryos in Arabidopsis. J. Exp. Bot. 2015, 66, 5029–5042. [Google Scholar] [CrossRef] [PubMed]
- Saini, S.; Sharma, I.; Kaur, N.; Pati, P.K. Auxin: A master regulator in plant root development. Plant Cell Rep. 2013, 32, 741–757. [Google Scholar] [CrossRef] [PubMed]
- Schaller, G.E.; Bishopp, A.; Kieber, J.J. The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 2015, 27, 44–63. [Google Scholar] [CrossRef] [PubMed]
- Skylar, A.; Wu, X. Regulation of Meristem Size by Cytokinin Signaling. J. Integr. Plant Biol. 2011, 53, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Teeples, M.; Lanctot, A.; Nemhauser, J.L. As above, so below: Auxin’s role in lateral organ development. Dev. Biol. 2016, 419, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Schmülling, T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009, 12, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Adamowski, M.; Friml, J. PIN-Dependent Auxin Transport: Action, Regulation, and Evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Argueso, C.T.; Raines, T.; Kieber, J.J. Cytokinin signaling and transcriptional networks. Curr. Opin. Plant Biol. 2010, 13, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Bennett, T. PIN proteins and the evolution of plant development. Trends Plant Sci. 2015, 20, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.; Sheen, J.; Müller, B. Cytokinin Signaling Networks. Annu. Rev. Plant Biol. 2012, 63, 353–380. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, H. Current aspects of auxin biosynthesis in plants. Biosci. Biotechnol. Biochem. 2016, 80, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Chen, J.; Yang, Z. Auxin regulation of cell polarity in plants. Curr. Opin. Plant Biol. 2015, 28, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Kieber, J.J.; Schaller, G.E. Cytokinins. Arabidopsis Book Am. Soc. Plant Biol. 2014, 12, e0168. [Google Scholar] [CrossRef] [PubMed]
- El-Showk, S.; Ruonala, R.; Helariutta, Y. Crossing paths: Cytokinin signalling and crosstalk. Development 2013, 140, 1373–1383. [Google Scholar] [CrossRef] [PubMed]
- Sehra, B.; Franks, R.G. Auxin and cytokinin act during gynoecial patterning and the development of ovules from the meristematic medial domain. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 555–571. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.-H.; Liu, Y.-B.; Zhang, X.-S. Auxin-cytokinin interaction regulates meristem development. Mol. Plant 2011, 4, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Vanstraelen, M.; Benková, E. Hormonal Interactions in the Regulation of Plant Development. Annu. Rev. Cell Dev. Biol. 2012, 28, 463–487. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Van Breusegem, F. ROS signaling: The new wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Tognetti, V.B.; Mühlenbock, P.; Van Breusegem, F. Stress homeostasis—The redox and auxin perspective. Plant Cell Environ. 2012, 35, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Auxin and the integration of environmental signals into plant root development. Ann. Bot. 2013, 112, 1655–1665. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, A.; Rathinasabapathi, B. Auxin and its transport play a role in plant tolerance to arsenite-induced oxidative stress in Arabidopsis thaliana. Plant Cell Environ. 2013, 36, 1838–1849. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.A.; Benkova, E. Cytokinin cross-talking during biotic and abiotic stress responses. Front. Plant Sci. 2013, 4, 451. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.-J.; Zhou, Y.-H.; Shi, K.; Zhou, J.; Foyer, C.H.; Yu, J.-Q. Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 2015, 66, 2839–2856. [Google Scholar] [CrossRef] [PubMed]
- Zwack, P.J.; Rashotte, A.M. Interactions between cytokinin signalling and abiotic stress responses. J. Exp. Bot. 2015, 66, 4863–4871. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Cerny, M.; Kuklova, A.; Hoehenwarter, W.; Fragner, L.; Novak, O.; Rotkova, G.; Jedelsky, P.L.; Zakova, K.; Smehilova, M.; Strnad, M.; et al. Proteome and metabolome profiling of cytokinin action in Arabidopsis identifying both distinct and similar responses to cytokinin down- and up-regulation. J. Exp. Bot. 2013, 64, 4193–4206. [Google Scholar] [CrossRef] [PubMed]
- Ljung, K.; Bhalerao, R.P.; Sandberg, G. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 2001, 28, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Mühlenbock, P.; Szechyńska-Hebda, M.; Płaszczyca, M.; Baudo, M.; Mateo, A.; Mullineaux, P.M.; Parker, J.E.; Karpińska, B.; Karpiński, S. Chloroplast Signaling and LESION SIMULATING DISEASE1 Regulate Crosstalk between Light Acclimation and Immunity in Arabidopsis. Plant Cell Online 2008, 20, 2339–2356. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Baek, D.; Park, H.C.; Chun, H.J.; Oh, D.-H.; Lee, M.K.; Cha, J.-Y.; Kim, W.-Y.; Kim, M.C.; Chung, W.S.; et al. Overexpression of Arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit. Mol. Plant 2013, 6, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Jung, J.-H.; Han, D.-Y.; Seo, P.J.; Park, W.J.; Park, C.-M. Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis. Planta 2012, 235, 923–938. [Google Scholar] [CrossRef] [PubMed]
- Park, H.C.; Cha, J.-Y.; Yun, D.-J. Roles of YUCCAs in auxin biosynthesis and drought stress responses in plants. Plant Signal. Behav. 2013, 8, e24495. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef] [PubMed]
- Sakata, T.; Oshino, T.; Miura, S.; Tomabechi, M.; Tsunaga, Y.; Higashitani, N.; Miyazawa, Y.; Takahashi, H.; Watanabe, M.; Higashitani, A. Auxins reverse plant male sterility caused by high temperatures. Proc. Natl. Acad. Sci. USA 2010, 107, 8569–8574. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.-Y.; Kim, W.-Y.; Kang, S.B.; Kim, J.I.; Baek, D.; Jung, I.J.; Kim, M.R.; Li, N.; Kim, H.-J.; Nakajima, M.; et al. A novel thiol-reductase activity of Arabidopsis YUC6 confers drought tolerance independently of auxin biosynthesis. Nat. Commun. 2015, 6, 8041. [Google Scholar] [CrossRef] [PubMed]
- Ke, Q.; Wang, Z.; Ji, C.Y.; Jeong, J.C.; Lee, H.-S.; Li, H.; Xu, B.; Deng, X.; Kwak, S.-S. Transgenic poplar expressing Arabidopsis YUCCA6 exhibits auxin-overproduction phenotypes and increased tolerance to abiotic stress. Plant Physiol. Biochem. 2015, 94, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Chen, L.; Ye, T.; Liu, X.; Ding, K.; Chan, Z. Modulation of auxin content in Arabidopsis confers improved drought stress resistance. Plant Physiol. Biochem. 2014, 82, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Woo, Y.-M.; Park, H.-J.; Su’udi, M.; Yang, J.-I.; Park, J.-J.; Back, K.; Park, Y.-M.; An, G. Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol. Biol. 2007, 65, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Nehnevajova, E.; Kollmer, I.; Novak, O.; Strnad, M.; Kramer, U.; Schmulling, T. Root-Specific Reduction of Cytokinin Causes Enhanced Root Growth, Drought Tolerance, and Leaf Mineral Enrichment in Arabidopsis and Tobacco. Plant Cell 2010, 22, 3905–3920. [Google Scholar] [CrossRef] [PubMed]
- Mok, D.W.; Mok, M.C. Cytokinin Metabolism and Action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 89–118. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Frebort, I.; Kowalska, M.; Hluska, T.; Frebortova, J.; Galuszka, P. Evolution of cytokinin biosynthesis and degradation. J. Exp. Bot. 2011, 62, 2431–2452. [Google Scholar] [CrossRef] [PubMed]
- Hare, P.D.; Cress, W.A.; van Staden, J. The involvement of cytokinins in plant responses to environmental stress. Plant Growth Regul. 1997, 23, 79–103. [Google Scholar] [CrossRef]
- Argueso, C.T.; Ferreira, F.J.; Kieber, J.J. Environmental perception avenues: The interaction of cytokinin and environmental response pathways. Plant Cell Environ. 2009, 32, 1147–1160. [Google Scholar] [CrossRef] [PubMed]
- Bano, A.; Hansen, H.; Dörffling, K.; Hahn, H. Changes in the contents of free and conjugated abscisic acid, phaseic acid and cytokinins in xylem sap of drought stressed sunflower plants. Phytochemistry 1994, 37, 345–347. [Google Scholar] [CrossRef]
- Shashidhar, V.R.; Prasad, T.G.; Sudharshan, L. Hormone signals from roots to shoots of sunflower (Helianthus annuus L.). Moderate soil drying increases delivery of abscisic acid and depresses delivery of cytokinins in xylem sap. Ann. Bot. 1996, 78, 151–155. [Google Scholar] [CrossRef]
- Alvarez, S.; Marsh, E.L.; Schroeder, S.G.; Schachtman, D.P. Metabolomic and proteomic changes in the xylem sap of maize under drought. Plant Cell Environ. 2008, 31, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Davies, W.J.; Kudoyarova, G.; Hartung, W. Long-distance ABA Signaling and Its Relation to Other Signaling Pathways in the Detection of Soil Drying and the Mediation of the Plant’s Response to Drought. J. Plant Growth Regul. 2005, 24, 285–295. [Google Scholar] [CrossRef]
- Hansen, H.; Dörffling, K. Root-derived trans-zeatin riboside and abscisic acid in drought-stressed and rewatered sunflower plants: Interaction in the control of leaf diffusive resistance? Funct. Plant Biol. 2003, 30, 365–375. [Google Scholar] [CrossRef]
- McDAVID, C.R.; Sagar, G.R.; Marshall, C. The Effect of Root Pruning and 6-Benzyl-Aminopurine on the Chlorophyll Content, 14CO2 Fixation and the Shoot/Root Ratio in Seedlings of Pisvm Sativum L. New Phytol. 1973, 72, 465–470. [Google Scholar] [CrossRef]
- Thomas, J.C.; McElwain, E.F.; Bohnert, H.J. Convergent Induction of Osmotic Stress-Responses 1. Plant Physiol. 1992, 100, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Lubovská, Z.; Dobrá, J.; Štorchová, H.; Wilhelmová, N.; Vanková, R. Cytokinin oxidase/dehydrogenase overexpression modifies antioxidant defense against heat, drought and their combination in Nicotiana tabacum plants. J. Plant Physiol. 2014, 171, 1625–1633. [Google Scholar] [CrossRef] [PubMed]
- Mackova, H.; Hronkova, M.; Dobra, J.; Tureckova, V.; Novak, O.; Lubovska, Z.; Motyka, V.; Haisel, D.; Hajek, T.; Prasil, I.T.; et al. Enhanced drought and heat stress tolerance of tobacco plants with ectopically enhanced cytokinin oxidase/dehydrogenase gene expression. J. Exp. Bot. 2013, 64, 2805–2815. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, R.; Watanabe, Y.; Fujita, Y.; Le, D.T.; Kojima, M.; Werner, T.; Vankova, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Kakimoto, T.; et al. Analysis of Cytokinin Mutants and Regulation of Cytokinin Metabolic Genes Reveals Important Regulatory Roles of Cytokinins in Drought, Salt and Abscisic Acid Responses, and Abscisic Acid Biosynthesis. Plant Cell 2011, 23, 2169–2183. [Google Scholar] [CrossRef] [PubMed]
- Pospíšilová, H.; Jiskrová, E.; Vojta, P.; Mrízová, K.; Kokáš, F.; Čudejková, M.M.; Bergougnoux, V.; Plíhal, O.; Klimešová, J.; Novák, O.; et al. Transgenic barley overexpressing a cytokinin dehydrogenase gene shows greater tolerance to drought stress. New Biotechnol. 2016, 33, 692–705. [Google Scholar] [CrossRef] [PubMed]
- Vojta, P.; Kokáš, F.; Husičková, A.; Grúz, J.; Bergougnoux, V.; Marchetti, C.F.; Jiskrová, E.; Ježilová, E.; Mik, V.; Ikeda, Y.; et al. Whole transcriptome analysis of transgenic barley with altered cytokinin homeostasis and increased tolerance to drought stress. New Biotechnol. 2016, 33, 676–691. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmulling, T. Cytokinin-Deficient Transgenic Arabidopsis Plants Show Multiple Developmental Alterations Indicating Opposite Functions of Cytokinins in the Regulation of Shoot and Root Meristem Activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef] [PubMed]
- Rivero, R.M.; Gimeno, J.; Deynze, A.V.; Walia, H.; Blumwald, E. Enhanced Cytokinin Synthesis in Tobacco Plants Expressing PSARK::IPT Prevents the Degradation of Photosynthetic Protein Complexes During Drought. Plant Cell Physiol. 2010, 51, 1929–1941. [Google Scholar] [CrossRef] [PubMed]
- Peleg, Z.; Reguera, M.; Tumimbang, E.; Walia, H.; Blumwald, E. Cytokinin-mediated source/sink modifications improve drought tolerance and increase grain yield in rice under water-stress. Plant Biotechnol. J. 2011, 9, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Gu, Q.; Zhang, J.; Sun, L.; Kuppu, S.; Zhang, Y.; Burow, M.; Payton, P.; Blumwald, E.; Zhang, H. Regulated Expression of an Isopentenyltransferase Gene (IPT) in Peanut Significantly Improves Drought Tolerance and Increases Yield Under Field Conditions. Plant Cell Physiol. 2011, 52, 1904–1914. [Google Scholar] [CrossRef] [PubMed]
- Kuppu, S.; Mishra, N.; Hu, R.; Sun, L.; Zhu, X.; Shen, G.; Blumwald, E.; Payton, P.; Zhang, H. Water-Deficit Inducible Expression of a Cytokinin Biosynthetic Gene IPT Improves Drought Tolerance in Cotton. PLoS ONE 2013, 8, e64190. [Google Scholar] [CrossRef] [PubMed]
- Décima Oneto, C.; Otegui, M.E.; Baroli, I.; Beznec, A.; Faccio, P.; Bossio, E.; Blumwald, E.; Lewi, D. Water deficit stress tolerance in maize conferred by expression of an isopentenyltransferase (IPT) gene driven by a stress- and maturation-induced promoter. J. Biotechnol. 2016, 220, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Rivero, R. M.; Kojima, M.; Gepstein, A.; Sakakibara, H.; Mittler, R.; Gepstein, S.; Blumwald, E. Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc. Natl. Acad. Sci. USA 2007, 104, 19631–19636. [Google Scholar] [CrossRef] [PubMed]
- Merewitz, E.B.; Gianfagna, T.; Huang, B. Photosynthesis, water use, and root viability under water stress as affected by expression of SAG12-ipt controlling cytokinin synthesis in Agrostis stolonifera. J. Exp. Bot. 2011, 62, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Burgess, P.; Zhang, X.; Huang, B. Enhancing cytokinin synthesis by overexpressing ipt alleviated drought inhibition of root growth through activating ROS-scavenging systems in Agrostis stolonifera. J. Exp. Bot. 2016, 67, 1979–1992. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shen, W.; Chan, Z.; Wu, Y. Endogenous Cytokinin Overproduction Modulates ROS Homeostasis and Decreases Salt Stress Resistance in Arabidopsis Thaliana. Front. Plant Sci. 2015, 6, 1004. [Google Scholar] [CrossRef] [PubMed]
- Žižková, E.; Dobrev, P.I.; Muhovski, Y.; Hošek, P.; Hoyerová, K.; Haisel, D.; Procházková, D.; Lutts, S.; Motyka, V.; Hichri, I. Tomato (Solanum lycopersicum L.) SlIPT3 and SlIPT4 isopentenyltransferases mediate salt stress response in tomato. BMC Plant Biol. 2015. [Google Scholar] [CrossRef]
- Tognetti, V.B.; Van Aken, O.; Morreel, K.; Vandenbroucke, K.; van de Cotte, B.; De Clercq, I.; Chiwocha, S.; Fenske, R.; Prinsen, E.; Boerjan, W.; et al. Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 2010, 22, 2660–2679. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, K.; Urano, K.; Yoshiwara, K.; Morishita, Y.; Sakurai, N.; Suzuki, H.; Kojima, M.; Sakakibara, H.; Shibata, D.; Saito, K.; et al. Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. Plant Physiol. 2014, 164, 1759–1771. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.K.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Narrowing down the targets for yield improvement in rice under normal and abiotic stress conditions via expression profiling of yield-related genes. Rice N. Y. 2012, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Cui, K.; Wang, W.; Li, Q.; Fahad, S.; Hu, Q.; Huang, J.; Nie, L.; Mohapatra, P.K.; Peng, S. Heat-Induced Cytokinin Transportation and Degradation Are Associated with Reduced Panicle Cytokinin Expression and Fewer Spikelets per Panicle in Rice. Front. Plant Sci. 2017, 8, 371. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Sahoo, K.K.; Tripathi, A.K.; Kumar, R.; Gupta, B.K.; Pareek, A.; Singla-Pareek, S.L. TKnockdown of an inflorescence meristem-specific cytokinin oxidase—OsCKX2 in rice reduces yield penalty under salinity stress condition. Plant Cell Environ. 2017. [Google Scholar] [CrossRef] [PubMed]
- Chang, Z.; Liu, Y.; Dong, H.; Teng, K.; Han, L.; Zhang, X. Effects of Cytokinin and Nitrogen on Drought Tolerance of Creeping Bentgrass. PLoS ONE 2016, 11, e0154005. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, B. Cytokinin Effects on Creeping Bentgrass Response to Heat Stress. Crop Sci. 2002, 42, 466–472. [Google Scholar] [CrossRef]
- Mýtinová, Z.; Motyka, V.; Haisel, D.; Gaudinová, A.; Lubovská, Z.; Wilhelmová, N. Effect of abiotic stresses on the activity of antioxidative enzymes and contents of phytohormones in wild type and AtCKX2 transgenic tobacco plants. Biol. Plant. 2010, 54, 461–470. [Google Scholar] [CrossRef]
- Skalák, J.; Černý, M.; Jedelský, P.; Dobrá, J.; Ge, E.; Novák, J.; Hronková, M.; Dobrev, P.; Vanková, R.; Brzobohatý, B. Stimulation of ipt overexpression as a tool to elucidate the role of cytokinins in high temperature responses of Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 2861–2873. [Google Scholar] [CrossRef] [PubMed]
- Zavaleta-Mancera, H.A.; López-Delgado, H.; Loza-Tavera, H.; Mora-Herrera, M.; Trevilla-García, C.; Vargas-Suárez, M.; Ougham, H. Cytokinin promotes catalase and ascorbate peroxidase activities and preserves the chloroplast integrity during dark-senescence. J. Plant Physiol. 2007, 164, 1572–1582. [Google Scholar] [CrossRef] [PubMed]
- Kawano, T. Roles of the reactive oxygen species-generating peroxidase reactions in plant defense and growth induction. Plant Cell Rep. 2003, 21, 829–837. [Google Scholar] [PubMed]
- Jansen, M.A.K.; van den Noort, R.E.; Tan, M.Y.A.; Prinsen, E.; Lagrimini, L.M.; Thorneley, R.N.F. Phenol-Oxidizing Peroxidases Contribute to the Protection of Plants from Ultraviolet Radiation Stress. Plant Physiol. 2001, 126, 1012–1023. [Google Scholar] [CrossRef] [PubMed]
- Vatulescu, A.D.; Fortunato, A.S.; Sá, M.C.; Amâncio, S.; Ricardo, C.P.P.; Jackson, P.A. Cloning and characterisation of a basic IAA oxidase associated with root induction in Vitis vinifera. Plant Physiol. Biochem. 2004, 42, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Cosio, C.; Vuillemin, L.; Meyer, M.D.; Kevers, C.; Penel, C.; Dunand, C. An anionic class III peroxidase from zucchini may regulate hypocotyl elongation through its auxin oxidase activity. Planta 2009, 229, 823–836. [Google Scholar] [CrossRef] [PubMed]
- Potters, G.; Pasternak, T.P.; Guisez, Y.; Jansen, M.A.K. Different stresses, similar morphogenic responses: integrating a plethora of pathways. Plant Cell Environ. 2009, 32, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Vanderauwera, S.; Zimmermann, P.; Rombauts, S.; Vandenabeele, S.; Langebartels, C.; Gruissem, W.; Inze, D.; Van Breusegem, F. Genome-Wide Analysis of Hydrogen Peroxide-Regulated Gene Expression in Arabidopsis Reveals a High Light-Induced Transcriptional Cluster Involved in Anthocyanin Biosynthesis. Plant Physiol. 2005, 139, 806–821. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Rubio-Moraga, A.; Trapero-Mozos, A.; Climent, M.F.L.; Gómez-Cadenas, A.; Gómez-Gómez, L. Ectopic expression of a stress-inducible glycosyltransferase from saffron enhances salt and oxidative stress tolerance in Arabidopsis while alters anchor root formation. Plant Sci. 2015, 234, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Ludwig-Müller, J. Auxin conjugates: Their role for plant development and in the evolution of land plants. J. Exp. Bot. 2011, 62, 1757–1773. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Wu, N.; Fu, J.; Wang, S.; Li, X.; Xiao, J.; Xiong, L. A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot. 2012, 63, 6467–6480. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-E.; Park, J.-Y.; Kim, Y.-S.; Staswick, P.E.; Jeon, J.; Yun, J.; Kim, S.-Y.; Kim, J.; Lee, Y.-H.; Park, C.-M. GH3-mediated Auxin Homeostasis Links Growth Regulation with Stress Adaptation Response in Arabidopsis. J. Biol. Chem. 2007, 282, 10036–10046. [Google Scholar] [CrossRef] [PubMed]
- Teichmann, T.; Bolu-Arianto, W.H.; Olbrich, A.; Langenfeld-Heyser, R.; Göbel, C.; Grzeganek, P.; Feussner, I.; Hänsch, R.; Polle, A. GH3::GUS reflects cell-specific developmental patterns and stress-induced changes in wood anatomy in the poplar stem. Tree Physiol. 2008, 28, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-W.; Li, C.-H.; Cao, J.; Zhang, Y.-C.; Zhang, S.-Q.; Xia, Y.-F.; Sun, D.-Y.; Sun, Y. Altered Architecture and Enhanced Drought Tolerance in Rice via the Down-Regulation of Indole-3-Acetic Acid by TLD1/OsGH3.13 Activation. Plant Physiol. 2009, 151, 1889–1901. [Google Scholar] [CrossRef] [PubMed]
- Junghans, U.; Polle, A.; Düchting, P.; Weiler, E.; Kuhlman, B.; Gruber, F.; Teichmann, T. Adaptation to high salinity in poplar involves changes in xylem anatomy and auxin physiology. Plant Cell Environ. 2006, 29, 1519–1531. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Jain, M.; Garg, R. Genome-wide analysis and expression profiling suggest diverse roles of GH3 genes during development and abiotic stress responses in legumes. Front. Plant Sci. 2015. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Yue, R.; Tao, S.; Yang, Y.; Zhang, L.; Xu, M.; Wang, H.; Shen, C. Genome-wide identification, expression analysis of auxin-responsive GH3 family genes in maize (Zea mays L.) under abiotic stresses. J. Integr. Plant Biol. 2015, 57, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, N.; Wang, H.; Kasahara, H.; Liu, J.; MacPherson, C.; Machida, Y.; Kamiya, Y.; Hannah, M.A.; Chua, N.-H. IAA-Ala Resistant3, an Evolutionarily Conserved Target of miR167, Mediates Arabidopsis Root Architecture Changes during High Osmotic Stress. Plant Cell 2012, 24, 3590–3602. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, M.; Ciarkowska, A.; Jakubowska, A. The auxin conjugate indole-3-acetyl-aspartate affects responses to cadmium and salt stress in Pisum sativum L. J. Plant Physiol. 2016, 191, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, B.; Dong, R.; Hou, B. AtUGT76C2, an Arabidopsis cytokinin glycosyltransferase is involved in drought stress adaptation. Plant Sci. 2015, 236, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Gidrol, X.; Lin, W.S.; Dégousée, N.; Yip, S.F.; Kush, A. Accumulation of Reactive Oxygen Species and Oxidation of Cytokinin in Germinating Soybean Seeds. Eur. J. Biochem. 1994, 224, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Peer, W.A.; Cheng, Y.; Murphy, A.S. Evidence of oxidative attenuation of auxin signalling. J. Exp. Bot. 2013, 64, 2629–2639. [Google Scholar] [CrossRef] [PubMed]
- Peer, W.A.; Murphy, A.S. Flavonoids and auxin transport: Modulators or regulators? Trends Plant Sci. 2007, 12, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Santelia, D.; Henrichs, S.; Vincenzetti, V.; Sauer, M.; Bigler, L.; Klein, M.; Bailly, A.; Lee, Y.; Friml, J.; Geisler, M.; et al. Flavonoids Redirect PIN-mediated Polar Auxin Fluxes during Root Gravitropic Responses. J. Biol. Chem. 2008, 283, 31218–31226. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, B.M.; Geisler, M.; Bigler, L.; Ringli, C. Flavonols Accumulate Asymmetrically and Affect Auxin Transport in Arabidopsis. Plant Physiol. 2011, 156, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Agati, G.; Brunetti, C.; Di Ferdinando, M.; Ferrini, F.; Pollastri, S.; Tattini, M. Functional roles of flavonoids in photoprotection: New evidence, lessons from the past. Plant Physiol. Biochem. 2013, 72, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.E.; Rashotte, A.M.; Murphy, A.S.; Normanly, J.; Tague, B.W.; Peer, W.A.; Taiz, L.; Muday, G.K. Flavonoids act as negative regulators of auxin transport in vivo in arabidopsis. Plant Physiol. 2001, 126, 524–535. [Google Scholar] [CrossRef] [PubMed]
- Buer, C.S.; Muday, G.K. The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 2004, 16, 1191–1205. [Google Scholar] [CrossRef] [PubMed]
- Peer, W.A.; Bandyopadhyay, A.; Blakeslee, J.J.; Makam, S.N.; Chen, R.J.; Masson, P.H.; Murphy, A.S. Variation in Expression and Protein Localization of the PIN Family of Auxin Efflux Facilitator Proteins in Flavonoid Mutants with Altered Auxin Transport in Arabidopsis thaliana. Plant Cell 2004, 16, 1898–1911. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, B.M.; Nodzyński, T.; Errafi, S.; Bucher, R.; Gupta, S.; Aryal, B.; Dobrev, P.; Bigler, L.; Geisler, M.; Zažímalová, E.; et al. Flavonol-induced changes in PIN2 polarity and auxin transport in the Arabidopsis thaliana rol1-2 mutant require phosphatase activity. Sci. Rep. 2017. [Google Scholar] [CrossRef] [PubMed]
- Buer, C.S.; Kordbacheh, F.; Truong, T.T.; Hocart, C.H.; Djordjevic, M.A. Alteration of flavonoid accumulation patterns in transparent testa mutants disturbs auxin transport, gravity responses, and imparts long-term effects on root and shoot architecture. Planta 2013, 238, 171–189. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, W.; De Smet, I.; Lewis, D.R.; Löfke, C.; Jansen, L.; Goeminne, G.; Vanden Bossche, R.; Karimi, M.; De Rybel, B.; Vanholme, B.; et al. Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc. Natl. Acad. Sci. USA 2012, 109, 1554–1559. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.P.; Nourizadeh, S.D.; Peer, W.A.; Xu, J.; Bandyopadhyay, A.; Murphy, A.S.; Goldsbrough, P.B. Arabidopsis AtGSTF2 is regulated by ethylene and auxin, and encodes a glutathione S-transferase that interacts with flavonoids. Plant J. 2003, 36, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Kokubo, T.; Ambe-Ono, Y.; Nakamura, M.; Ishida, H.; Yamakawa, T.; Kodama, T. Promotive effect of auxins on UDP-glucose: Flavonol glucosyltransferase activity in Vitis sp. cell cultures. J. Biosci. Bioeng. 2001, 91, 564–569. [Google Scholar] [CrossRef]
- Silva-Navas, J.; Moreno-Risueno, M.A.; Manzano, C.; Téllez-Robledo, B.; Navarro-Neila, S.; Carrasco, V.; Pollmann, S.; Gallego, F.J.; del Pozo, J.C. Flavonols Mediate Root Phototropism and Growth through Regulation of Proliferation-to-Differentiation Transition. Plant Cell 2016, 28, 1372–1387. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, T.; Rudas, V.; Potters, G.; Jansen, M.A.K. Morphogenic effects of abiotic stress: Reorientation of growth in Arabidopsis thaliana seedlings. Environ. Exp. Bot. 2005, 53, 299–314. [Google Scholar] [CrossRef]
- Jiang, Y.; Deyholos, M.K. Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol. 2006, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, R.-J.; Han, T.-T.; Cai, W.; Fu, Z.-W.; Lu, Y.-T. Salt Stress Reduces Root Meristem Size by Nitric Oxide-Mediated Modulation of Auxin Accumulation and Signaling in Arabidopsis. Plant Physiol. 2015, 168, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Zhang, W.; Hu, H.; Li, B.; Wang, Y.; Zhao, Y.; Li, K.; Liu, M.; Li, X. Salt Modulates Gravity Signaling Pathway to Regulate Growth Direction of Primary Roots in Arabidopsis. Plant Physiol. 2008, 146, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Yue, R.; Tie, S.; Sun, T.; Zhang, L.; Yang, Y.; Qi, J.; Yan, S.; Han, X.; Wang, H.; Shen, C. Genome-Wide Identification and Expression Profiling Analysis of ZmPIN, ZmPILS, ZmLAX and ZmABCB Auxin Transporter Gene Families in Maize (Zea mays L.) under Various Abiotic Stresses. PLoS ONE 2015, 10, e0118751. [Google Scholar] [CrossRef] [PubMed]
- Geldner, N.; Anders, N.; Wolters, H.; Keicher, J.; Kornberger, W.; Muller, P.; Delbarre, A.; Ueda, T.; Nakano, A.; Jürgens, G. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 2003, 112, 219–230. [Google Scholar] [CrossRef]
- Kleine-Vehn, J.; Łangowski, Ł.; Wiśniewska, J.; Dhonukshe, P.; Brewer, P.B.; Friml, J. Cellular and Molecular Requirements for Polar PIN Targeting and Transcytosis in Plants. Mol. Plant 2008, 1, 1056–1066. [Google Scholar] [CrossRef] [PubMed]
- Benková, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertová, D.; Jürgens, G.; Friml, J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef]
- Friml, J.; Wiśniewska, J.; Benková, E.; Mendgen, K.; Palme, K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 2002, 415, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jürgens, G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, D.; Pesce, E.-R.; Stieger, P.; Mandel, T.; Baltensperger, K.; Bennett, M.; Traas, J.; Friml, J.; Kuhlemeier, C. Regulation of phyllotaxis by polar auxin transport. Nature 2003, 426, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Heisler, M.G.; Ohno, C.; Das, P.; Sieber, P.; Reddy, G.V.; Long, J.A.; Meyerowitz, E.M. Patterns of Auxin Transport and Gene Expression during Primordium Development Revealed by Live Imaging of the Arabidopsis Inflorescence Meristem. Curr. Biol. 2005, 15, 1899–1911. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, K.; Uemura, M.; Tsurumi, S.; Rahman, A. Auxin Response in Arabidopsis under Cold Stress: Underlying Molecular Mechanisms. Plant Cell 2009, 21, 3823–3838. [Google Scholar] [CrossRef] [PubMed]
- Zwiewka, M.; Nodzyński, T.; Robert, S.; Vanneste, S.; Friml, J. Osmotic Stress Modulates the Balance between Exocytosis and Clathrin-Mediated Endocytosis in Arabidopsis thaliana. Mol. Plant 2015, 8, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Galvan-Ampudia, C.S.; Julkowska, M.M.; Darwish, E.; Gandullo, J.; Korver, R.A.; Brunoud, G.; Haring, M.A.; Munnik, T.; Vernoux, T.; Testerink, C. Halotropism Is a Response of Plant Roots to Avoid a Saline Environment. Curr. Biol. 2013, 23, 2044–2050. [Google Scholar] [CrossRef] [PubMed]
- Bishopp, A.; Help, H.; El-Showk, S.; Weijers, D.; Scheres, B.; Friml, J.; Benková, E.; Mähönen, A.P.; Helariutta, Y. A Mutually Inhibitory Interaction between Auxin and Cytokinin Specifies Vascular Pattern in Roots. Curr. Biol. 2011, 21, 917–926. [Google Scholar] [CrossRef] [PubMed]
- Gillissen, B.; Bürkle, L.; André, B.; Kühn, C.; Rentsch, D.; Brandl, B.; Frommer, W.B. A New Family of High-Affinity Transporters for Adenine, Cytosine, and Purine Derivatives in Arabidopsis. Plant Cell 2000, 12, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Kudo, T.; Kiba, T.; Sakakibara, H. Metabolism and Long-distance Translocation of Cytokinins. J. Integr. Plant Biol. 2010, 52, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Bürkle, L.; Cedzich, A.; Döpke, C.; Stransky, H.; Okumoto, S.; Gillissen, B.; Kühn, C.; Frommer, W.B. Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. Plant J. 2003, 34, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Hirose, N.; Takei, K.; Kuroha, T.; Kamada-Nobusada, T.; Hayashi, H.; Sakakibara, H. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 2007, 59, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Bishopp, A.; Lehesranta, S.; Vatén, A.; Help, H.; El-Showk, S.; Scheres, B.; Helariutta, K.; Mähönen, A.P.; Sakakibara, H.; Helariutta, Y. Phloem-Transported Cytokinin Regulates Polar Auxin Transport and Maintains Vascular Pattern in the Root Meristem. Curr. Biol. 2011, 21, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Ko, D.; Kang, J.; Kiba, T.; Park, J.; Kojima, M.; Do, J.; Kim, K.Y.; Kwon, M.; Endler, A.; Song, W.-Y.; et al. Arabidopsis ABCG14 is essential for the root-to-shoot translocation of cytokinin. Proc. Natl. Acad. Sci. USA 2014, 111, 7150–7155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Novak, O.; Wei, Z.; Gou, M.; Zhang, X.; Yu, Y.; Yang, H.; Cai, Y.; Strnad, M.; Liu, C.-J. Arabidopsis ABCG14 protein controls the acropetal translocation of root-synthesized cytokinins. Nat. Commun. 2014, 5, 3274. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Murfett, J.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell Online 1997, 9, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
- Shu, W.; Liu, Y.; Guo, Y.; Zhou, H.; Zhang, J.; Zhao, S.; Lu, M. A Populus TIR1 gene family survey reveals differential expression patterns and responses to 1-naphthaleneacetic acid and stress treatments. Front. Plant Sci. 2015, 6, 719. [Google Scholar] [PubMed]
- Blomster, T.; Salojarvi, J.; Sipari, N.; Brosche, M.; Ahlfors, R.; Keinanen, M.; Overmyer, K.; Kangasjarvi, J. Apoplastic Reactive Oxygen Species Transiently Decrease Auxin Signaling and Cause Stress-Induced Morphogenic Response in Arabidopsis. Plant Physiol. 2011, 157, 1866–1883. [Google Scholar] [CrossRef] [PubMed]
- Farcot, E.; Lavedrine, C.; Vernoux, T. A modular analysis of the auxin signalling network. PLoS ONE 2015, 10, e0122231. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, M.J.; Terrile, M.C.; Bartoli, C.G.; D’Ippólito, S.; Casalongué, C.A. Auxin signaling participates in the adaptative response against oxidative stress and salinity by interacting with redox metabolism in Arabidopsis. Plant Mol. Biol. 2010, 74, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.V.; Le, D.T.; Nishiyama, R.; Watanabe, Y.; Sulieman, S.; Tran, U.T.; Mochida, K.; Dong, N.V.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; et al. The auxin response factor transcription factor family in soybean: Genome-wide identification and expression analyses during development and water stress. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2013, 20, 511–524. [Google Scholar]
- Jain, M.; Khurana, J.P. Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS J. 2009, 276, 3148–3162. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Bai, Y.; Shen, C.; Wu, Y.; Zhang, S.; Jiang, D.; Guilfoyle, T.J.; Chen, M.; Qi, Y. Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct. Integr. Genom. 2010, 10, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Lee, D.-K.; Choi, Y.D.; Kim, J.-K. OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci. 2015, 236, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Matsui, A.; Ishida, J.; Morosawa, T.; Mochizuki, Y.; Kaminuma, E.; Endo, T.A.; Okamoto, M.; Nambara, E.; Nakajima, M.; Kawashima, M.; et al. Arabidopsis Transcriptome Analysis under Drought, Cold, High-Salinity and ABA Treatment Conditions using a Tiling Array. Plant Cell Physiol. 2008, 49, 1135–1149. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Xu, L.; Wang, Y.; Yu, R.; Zhu, X.; Luo, X.; Gong, Y.; Wang, R.; Limera, C.; Zhang, K.; et al. Identification of novel and salt-responsive miRNAs to explore miRNA-mediated regulatory network of salt stress response in radish (Raphanus sativus L.). BMC Genom. 2015, 16, 197. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-H.; Tian, X.; Li, Y.-J.; Wu, C.-A.; Zheng, C.-C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 2008, 14, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Zürcher, E.; Müller, B. Cytokinin Synthesis, Signaling, and Function—Advances and New Insights. Int. Rev. Cell Mol. Biol. 2016, 324, 1–38. [Google Scholar] [PubMed]
- Argyros, R.D.; Mathews, D.E.; Chiang, Y.-H.; Palmer, C.M.; Thibault, D.M.; Etheridge, N.; Argyros, D.A.; Mason, M.G.; Kieber, J.J.; Schaller, G.E. Type B Response Regulators of Arabidopsis Play Key Roles in Cytokinin Signaling and Plant Development. Plant Cell Online 2008, 20, 2102–2116. [Google Scholar] [CrossRef] [PubMed]
- Sakai, H.; Aoyama, T.; Bono, H.; Oka, A. Two-Component Response Regulators from Arabidopsis thaliana Contain a Putative DNA-Binding Motif. Plant Cell Physiol. 1998, 39, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Sakai, H.; Aoyama, T.; Oka, A. Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. Cell Mol. Biol. 2000, 24, 703–711. [Google Scholar] [CrossRef]
- Sakai, H.; Honma, T.; Aoyama, T.; Sato, S.; Kato, T.; Tabata, S.; Oka, A. ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 2001, 294, 1519–1521. [Google Scholar] [CrossRef] [PubMed]
- Brandstatter, I.; Kieber, J.J. Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 1998, 10, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, I.B.; Deruère, J.; Kieber, J.J. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 2000, 124, 1706–1717. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.; Sheen, J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 2001, 413, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.-S.P.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 20623–20628. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.N.; Verslues, P.E. Stress physiology functions of the Arabidopsis histidine kinase cytokinin receptors. Physiol. Plant. 2015, 154, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Kim, N.Y.; Kim, S.; Kang, N.Y.; Novak, O.; Ku, S.-J.; Cho, C.; Lee, D.J.; Lee, E.-J.; Strnad, M.; et al. A Subset of Cytokinin Two-component Signaling System Plays a Role in Cold Temperature Stress Response in Arabidopsis. J. Biol. Chem. 2010, 285, 23371–23386. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Kim, J. Arabidopsis Response Regulator1 and Arabidopsis Histidine Phosphotransfer Protein2 (AHP2), AHP3, and AHP5 Function in Cold Signaling. Plant Physiol. 2013, 161, 408–424. [Google Scholar] [CrossRef] [PubMed]
- Kang, N.Y.; Cho, C.; Kim, J. Inducible Expression of Arabidopsis Response Regulator 22 (ARR22), a Type-C ARR, in Transgenic Arabidopsis Enhances Drought and Freezing Tolerance. PLoS ONE 2013. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, A.; Clabaugh, I.; To, J.P.; Maxwell, B.B.; Chiang, Y.-H.; Schaller, G.E.; Loraine, A.; Kieber, J.J. Identification of Cytokinin-Responsive Genes Using Microarray Meta-Analysis and RNA-Seq in Arabidopsis. Plant Physiol. 2013, 162, 272–294. [Google Scholar] [CrossRef] [PubMed]
- Brenner, W.G.; Schmulling, T. Transcript profiling of cytokinin action in Arabidopsis roots and shoots discovers largely similar but also organ-specific responses. BMC Plant Biol. 2012, 12, 112. [Google Scholar] [CrossRef] [PubMed]
- Kocsy, G.; Tari, I.; Vanková, R.; Zechmann, B.; Gulyás, Z.; Poór, P.; Galiba, G. Redox control of plant growth and development. Plant Sci. 2013, 211, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Reguera, M.; Peleg, Z.; Abdel-Tawab, Y.M.; Tumimbang, E.B.; Delatorre, C.A.; Blumwald, E. Stress-Induced Cytokinin Synthesis Increases Drought Tolerance through the Coordinated Regulation of Carbon and Nitrogen Assimilation in Rice. Plant Physiol. 2013, 163, 1609–1622. [Google Scholar] [CrossRef] [PubMed]
- Kang, N.Y.; Cho, C.; Kim, N.Y.; Kim, J. Cytokinin receptor-dependent and receptor-independent pathways in the dehydration response of Arabidopsis thaliana. J. Plant Physiol. 2012, 169, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Dello Ioio, R.; Linhares, F.S.; Scacchi, E.; Casamitjana-Martinez, E.; Heidstra, R.; Costantino, P.; Sabatini, S. Cytokinins Determine Arabidopsis Root-Meristem Size by Controlling Cell Differentiation. Curr. Biol. 2007, 17, 678–682. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, R.; Watanabe, Y.; Leyva-Gonzalez, M.A.; Van Ha, C.; Fujita, Y.; Tanaka, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Herrera-Estrella, L.; et al. Arabidopsis AHP2, AHP3, and AHP5 histidine phosphotransfer proteins function as redundant negative regulators of drought stress response. Proc. Natl. Acad. Sci. USA 2013, 110, 4840–4845. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, Q.; Wu, J.; Zhang, L.; Jiao, X.; Zhang, S.; Zhang, Z.; Sun, D.; Lu, T.; Sun, Y. Two Rice Authentic Histidine Phosphotransfer Proteins, OsAHP1 and OsAHP2, Mediate Cytokinin Signaling and Stress Responses in Rice. Plant Physiol. 2014, 165, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Mason, M.G.; Jha, D.; Salt, D.E.; Tester, M.; Hill, K.; Kieber, J.J.; Eric Schaller, G. Type-B response regulators ARR1 and ARR12 regulate expression of AtHKT1;1 and accumulation of sodium in Arabidopsis shoots. Plant J. 2010, 64, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Rashotte, A.M.; Mason, M.G.; Hutchison, C.E.; Ferreira, F.J.; Schaller, G.E.; Kieber, J.J. A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. USA 2006, 103, 11081–11085. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Gupta, S.; Rashotte, A.M. Characterization of two tomato AP2/ERF genes, SlCRF1 and SlCRF2 in hormone and stress responses. Plant Cell Rep. 2014, 33, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Inzé, A.; Vanderauwera, S.; Hoeberichts, F.A.; Vandorpe, M.; van Gaever, T.; Van Breusegem, F. A subcellular localization compendium of hydrogen peroxide-induced proteins. Plant Cell Environ. 2012, 35, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Zwack, P.J.; Robinson, B.R.; Risley, M.G.; Rashotte, A.M. Cytokinin Response Factor 6 Negatively Regulates Leaf Senescence and is Induced in Response to Cytokinin and Numerous Abiotic Stresses. Plant Cell Physiol. 2013, 54, 971–981. [Google Scholar] [CrossRef] [PubMed]
- Zwack, P.J.; De Clercq, I.; Howton, T.C.; Hallmark, H.T.; Hurny, A.; Keshishian, E.A.; Parish, A.M.; Benkova, E.; Mukhtar, M.S.; Van Breusegem, F.; et al. Cytokinin Response Factor 6 Represses Cytokinin-Associated Genes during Oxidative Stress. Plant Physiol. 2016, 172, 1249–1258. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Rashotte, A.M. Expression patterns and regulation of SlCRF3 and SlCRF5 in response to cytokinin and abiotic stresses in tomato (Solanum lycopersicum). J. Plant Physiol. 2014, 171, 349–358. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, I.; Vermeirssen, V.; van Aken, O.; Vandepoele, K.; Murcha, M.W.; Law, S.R.; Inzé, A.; Ng, S.; Ivanova, A.; Rombaut, D.; et al. The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 2013, 25, 3472–3490. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; Ivanova, A.; Duncan, O.; Law, S.R.; Van Aken, O.; de Clercq, I.; Wang, Y.; Carrie, C.; Xu, L.; Kmiec, B.; et al. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 2013, 25, 3450–3471. [Google Scholar] [CrossRef] [PubMed]
- Zwack, P.J.; Compton, M.A.; Adams, C.I.; Rashotte, A.M. Cytokinin response factor 4 (CRF4) is induced by cold and involved in freezing tolerance. Plant Cell Rep. 2015, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wang, C.; Chen, Q.; Chen, H.; Ren, B.; Li, X.; Zuo, J. S-nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nat. Commun. 2013, 4, 1529. [Google Scholar] [CrossRef] [PubMed]
- Terrile, M.C.; París, R.; Calderón-Villalobos, L.I.A.; Iglesias, M.J.; Lamattina, L.; Estelle, M.; Casalongué, C.A. Nitric Oxide Influences Auxin Signaling through S-nitrosylation of the Arabidopsis Transport Inhibitor Response1 Auxin Receptor. Plant J. 2012, 70, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.; Ljung, K. Auxin and cytokinin regulate each other’s levels via a metabolic feedback loop. Plant Signal. Behav. 2011, 6, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, K.D. How many ways are there to make a root? Curr. Opin. Plant Biol. 2016, 34, 61–67. [Google Scholar] [CrossRef] [PubMed]
- De Rybel, B.; Mähönen, A.P.; Helariutta, Y.; Weijers, D. Plant vascular development: From early specification to differentiation. Nat. Rev. Mol. Cell Biol. 2016, 17, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Naseem, M.; Kaltdorf, M.; Dandekar, T. The nexus between growth and defence signalling: Auxin and cytokinin modulate plant immune response pathways. J. Exp. Bot. 2015, 66, 4885–4896. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, K.; Matsumoto-Kitano, M.; Kakimoto, T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: Tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. Cell Mol. Biol. 2004, 37, 128–138. [Google Scholar] [CrossRef]
- Dello Ioio, R.; Nakamura, K.; Moubayidin, L.; Perilli, S.; Taniguchi, M.; Morita, M.T.; Aoyama, T.; Costantino, P.; Sabatini, S. A genetic framework for the control of cell division and differentiation in the root meristem. Science 2008, 322, 1380–1384. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Takei, K.; Kojima, M.; Sakakibara, H.; Mori, H. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 2006, 45, 1028–1036. [Google Scholar] [CrossRef] [PubMed]
- Nordström, A.; Tarkowski, P.; Tarkowska, D.; Norbaek, R.; Astot, C.; Dolezal, K.; Sandberg, G. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: A factor of potential importance for auxin-cytokinin-regulated development. Proc. Natl. Acad. Sci. USA 2004, 101, 8039–8044. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Köllmer, I.; Bartrina, I.; Holst, K.; Schmülling, T. New Insights into the Biology of Cytokinin Degradation. Plant Biol. 2006, 8, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Carabelli, M.; Possenti, M.; Sessa, G.; Ciolfi, A.; Sassi, M.; Morelli, G.; Ruberti, I. Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev. 2007, 21, 1863–1868. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.; Gunnerås, S.A.; Petersson, S.V.; Tarkowski, P.; Graham, N.; May, S.; Dolezal, K.; Sandberg, G.; Ljung, K. Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell 2010, 22, 2956–2969. [Google Scholar] [CrossRef] [PubMed]
- Polanská, L.; Vicánková, A.; Nováková, M.; Malbeck, J.; Dobrev, P.I.; Brzobohaty, B.; Vanková, R.; Machácková, I. Altered cytokinin metabolism affects cytokinin, auxin, and abscisic acid contents in leaves and chloroplasts, and chloroplast ultrastructure in transgenic tobacco. J. Exp. Bot. 2007, 58, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Müller, B.; Sheen, J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 2008, 453, 1094–1097. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Andersen, S.U.; Ljung, K.; Dolezal, K.; Miotk, A.; Schultheiss, S.J.; Lohmann, J.U. Hormonal control of the shoot stem-cell niche. Nature 2010, 465, 1089–1092. [Google Scholar] [CrossRef] [PubMed]
- Moubayidin, L.; Perilli, S.; Dello Ioio, R.; Di Mambro, R.; Costantino, P.; Sabatini, S. The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr. Biol. 2010, 20, 1138–1143. [Google Scholar] [CrossRef] [PubMed]
- Pernisová, M.; Klíma, P.; Horák, J.; Válková, M.; Malbeck, J.; Souček, P.; Reichman, P.; Hoyerová, K.; Dubová, J.; Friml, J.; et al. Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc. Natl. Acad. Sci. USA 2009, 106, 3609–3614. [Google Scholar] [CrossRef] [PubMed]
- Marhavý, P.; Bielach, A.; Abas, L.; Abuzeineh, A.; Duclercq, J.; Tanaka, H.; Pařezová, M.; Petrášek, J.; Friml, J.; Kleine-Vehn, J.; et al. Cytokinin Modulates Endocytic Trafficking of PIN1 Auxin Efflux Carrier to Control Plant Organogenesis. Dev. Cell 2011, 21, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Rŭžička, K.; Šimášková, M.; Duclercq, J.; Petrášek, J.; Zažímalová, E.; Simon, S.; Friml, J.; Van Montagu, M.C.; Benková, E. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl. Acad. Sci. USA 2009, 106, 4284–4289. [Google Scholar] [CrossRef] [PubMed]
- Moreira, S.; Bishopp, A.; Carvalho, H.; Campilho, A. AHP6 Inhibits Cytokinin Signaling to Regulate the Orientation of Pericycle Cell Division during Lateral Root Initiation. PLoS ONE 2013. [Google Scholar] [CrossRef] [PubMed]
- Kuppusamy, K.T.; Walcher, C.L.; Nemhauser, J.L. Cross-regulatory mechanisms in hormone signaling. Plant Mol. Biol. 2009, 69, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Ramireddy, E.; Chang, L.; Schmülling, T. Cytokinin as a mediator for regulating root system architecture in response to environmental cues. Plant Signal. Behav. 2014. [Google Scholar] [CrossRef]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator V3: A Reference Expression Database for the Meta-Analysis of Transcriptomes. Adv. Bioinform. 2008, 2008, e420747. [Google Scholar] [CrossRef] [PubMed]
- Bennett, T.; Hines, G.; van Rongen, M.; Waldie, T.; Sawchuk, M.G.; Scarpella, E.; Ljung, K.; Leyser, O. Connective Auxin Transport in the Shoot Facilitates Communication between Shoot Apices. PLoS Biol. 2016, 14, e1002446. [Google Scholar] [CrossRef] [PubMed]
- Bainbridge, K.; Guyomarc’h, S.; Bayer, E.; Swarup, R.; Bennett, M.; Mandel, T.; Kuhlemeier, C. Auxin influx carriers stabilize phyllotactic patterning. Genes Dev. 2008, 22, 810–823. [Google Scholar] [CrossRef] [PubMed]
- Revalska, M.; Vassileva, V.; Zechirov, G.; Iantcheva, A. Is the auxin influx carrier LAX3 essential for plant growth and development in the model plants Medicago truncatula, Lotus japonicus and Arabidopsis thaliana? Biotechnol. Biotechnol. Equip. 2015, 29, 786–797. [Google Scholar] [CrossRef]
- Wolters, H.; Anders, N.; Geldner, N.; Gavidia, R.; Jürgens, G. Coordination of apical and basal embryo development revealed by tissue-specific GNOM functions. Development 2011, 138, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Gray, W.M. SAUR Proteins as Effectors of Hormonal and Environmental Signals in Plant Growth. Mol. Plant 2015, 8, 1153. [Google Scholar] [CrossRef] [PubMed]
- Chae, K.; Isaacs, C.G.; Reeves, P.H.; Maloney, G.S.; Muday, G.K.; Nagpal, P.; Reed, J.W. Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J. 2012, 71, 684–697. [Google Scholar] [CrossRef] [PubMed]
- Spartz, A.K.; Ren, H.; Park, M.Y.; Grandt, K.N.; Lee, S.H.; Murphy, A.S.; Sussman, M.R.; Overvoorde, P.J.; Gray, W.M. SAUR Inhibition of PP2C-D Phosphatases Activates Plasma Membrane H+-ATPases to Promote Cell Expansion in Arabidopsis. Plant Cell 2014, 26, 2129–2142. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Richardson, T.; Chai, S.; McIntyre, C.L.; Rae, A.L.; Xue, G.-P. Drought-Up-Regulated TaNAC69-1 is a Transcriptional Repressor of TaSHY2 and TaIAA7, and Enhances Root Length and Biomass in Wheat. Plant Cell Physiol. 2016, 57, 2076–2090. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Bai, M.-Y.; Kim, J.-G.; Wang, T.; Oh, E.; Chen, L.; Park, C.H.; Son, S.-H.; Kim, S.-K.; Mudgett, M.B.; et al. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell 2014, 26, 828–841. [Google Scholar] [CrossRef] [PubMed]
- Benekos, K.; Kissoudis, C.; Nianiou-Obeidat, I.; Labrou, N.; Madesis, P.; Kalamaki, M.; Makris, A.; Tsaftaris, A. Overexpression of a specific soybean GmGSTU4 isoenzyme improves diphenyl ether and chloroacetanilide herbicide tolerance of transgenic tobacco plants. J. Biotechnol. 2010, 150, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Jha, Y.; Subramanian, R.B.; Patel, S. Combination of endophytic and rhizospheric plant growth promoting rhizobacteria in Oryza sativa shows higher accumulation of osmoprotectant against saline stress. Acta Physiol. Plant. 2011, 33, 797–802. [Google Scholar] [CrossRef]
- Roxas, V.P.; Smith, R.K.; Allen, E.R.; Allen, R.D. Overexpression of glutathione S-transferase/glutathioneperoxidase enhances the growth of transgenic tobacco seedlings during stress. Nat. Biotechnol. 1997, 15, 988–991. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Sahoo, A.; Devendran, R.; Jain, M. Over-Expression of a Rice Tau Class Glutathione S-Transferase Gene Improves Tolerance to Salinity and Oxidative Stresses in Arabidopsis. PLoS ONE 2014. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, V.; Patel, M.K.; Chaturvedi, A.K.; Mishra, A.; Jha, B. Functional Characterization of the Tau Class Glutathione-S-Transferases Gene (SbGSTU) Promoter of Salicornia brachiata under Salinity and Osmotic Stress. PLoS ONE 2016. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Vivancos, P.; de Simone, A.; Kiddle, G.; Foyer, C.H. Glutathione—Linking cell proliferation to oxidative stress. Free Radic. Biol. Med. 2015, 89, 1154–1164. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Ascorbate and Glutathione: The Heart of the Redox Hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2012, 35, 454–484. [Google Scholar] [CrossRef] [PubMed]
- Passaia, G.; Queval, G.; Bai, J.; Margis-Pinheiro, M.; Foyer, C.H. The effects of redox controls mediated by glutathione peroxidases on root architecture in Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 1403–1413. [Google Scholar] [CrossRef] [PubMed]
- Remy, E.; Cabrito, T.R.; Baster, P.; Batista, R.A.; Teixeira, M.C.; Friml, J.; Sa-Correia, I.; Duque, P. A Major Facilitator Superfamily Transporter Plays a Dual Role in Polar Auxin Transport and Drought Stress Tolerance in Arabidopsis. Plant Cell 2013, 25, 901–926. [Google Scholar] [CrossRef] [PubMed]
- Kamimoto, Y.; Terasaka, K.; Hamamoto, M.; Takanashi, K.; Fukuda, S.; Shitan, N.; Sugiyama, A.; Suzuki, H.; Shibata, D.; Wang, B.; et al. Arabidopsis ABCB21 is a Facultative Auxin Importer/Exporter Regulated by Cytoplasmic Auxin Concentration. Plant Cell Physiol. 2012, 53, 2090–2100. [Google Scholar] [CrossRef] [PubMed]
- Terasaka, K.; Blakeslee, J.J.; Titapiwatanakun, B.; Peer, W.A.; Bandyopadhyay, A.; Makam, S.N.; Lee, O.R.; Richards, E.L.; Murphy, A.S.; Sato, F.; et al. PGP4, an ATP Binding Cassette P-Glycoprotein, Catalyzes Auxin Transport in Arabidopsis thaliana Roots. Plant Cell 2005, 17, 2922–2939. [Google Scholar] [CrossRef] [PubMed]
- Barbez, E.; Kubeš, M.; Rolčík, J.; Béziat, C.; Pěnčík, A.; Wang, B.; Rosquete, M.R.; Zhu, J.; Dobrev, P.I.; Lee, Y.; et al. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 2012, 485, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, S.; Hishiyama, S.; Jikumaru, Y.; Hanada, A.; Nishimura, T.; Koshiba, T.; Zhao, Y.; Kamiya, Y.; Kasahara, H. Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 5430–5435. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Auxin Biosynthesis. Arabidopsis Book Am. Soc. Plant Biol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Mellor, N.; Band, L.R.; Pěnčík, A.; Novák, O.; Rashed, A.; Holman, T.; Wilson, M.H.; Voß, U.; Bishopp, A.; King, J.R.; et al. Dynamic regulation of auxin oxidase and conjugating enzymes AtDAO1 and GH3 modulates auxin homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, 11022–11027. [Google Scholar] [CrossRef] [PubMed]
- Bou-Torrent, J.; Salla-Martret, M.; Brandt, R.; Musielak, T.; Palauqui, J.-C.; Martínez-García, J.F.; Wenkel, S. ATHB4 and HAT3, two class II HD-ZIP transcription factors, control leaf development in Arabidopsis. Plant Signal. Behav. 2012, 7, 1382–1387. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Xie, Q.; Chua, N.-H. The Arabidopsis Auxin-Inducible Gene ARGOS Controls Lateral Organ Size. Plant Cell 2003, 15, 1951. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Habben, J.E.; Archibald, R.L.; Drummond, B.J.; Chamberlin, M.A.; Williams, R.W.; Lafitte, H.R.; Weers, B.P. Overexpression of ARGOS Genes Modifies Plant Sensitivity to Ethylene, Leading to Improved Drought Tolerance in Both Arabidopsis and Maize. Plant Physiol. 2015, 169, 266–282. [Google Scholar] [CrossRef] [PubMed]
- Rawat, R.; Schwartz, J.; Jones, M.A.; Sairanen, I.; Cheng, Y.; Andersson, C.R.; Zhao, Y.; Ljung, K.; Harmer, S.L. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc. Natl. Acad. Sci. USA 2009, 106, 16883–16888. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Chiang, Y.-H.; Kieber, J.J.; Schaller, G.E. SCFKMD controls cytokinin signaling by regulating the degradation of type-B response regulators. Proc. Natl. Acad. Sci. USA 2013, 110, 10028. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gou, M.; Guo, C.; Yang, H.; Liu, C.-J. Down-Regulation of Kelch Domain-Containing F-Box Protein in Arabidopsis Enhances the Production of (Poly) phenols and Tolerance to Ultraviolet Radiation. Plant Physiol. 2015, 167, 337. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bielach, A.; Hrtyan, M.; Tognetti, V.B. Plants under Stress: Involvement of Auxin and Cytokinin. Int. J. Mol. Sci. 2017, 18, 1427. https://doi.org/10.3390/ijms18071427
Bielach A, Hrtyan M, Tognetti VB. Plants under Stress: Involvement of Auxin and Cytokinin. International Journal of Molecular Sciences. 2017; 18(7):1427. https://doi.org/10.3390/ijms18071427
Chicago/Turabian StyleBielach, Agnieszka, Monika Hrtyan, and Vanesa B. Tognetti. 2017. "Plants under Stress: Involvement of Auxin and Cytokinin" International Journal of Molecular Sciences 18, no. 7: 1427. https://doi.org/10.3390/ijms18071427
APA StyleBielach, A., Hrtyan, M., & Tognetti, V. B. (2017). Plants under Stress: Involvement of Auxin and Cytokinin. International Journal of Molecular Sciences, 18(7), 1427. https://doi.org/10.3390/ijms18071427