Novel Molecular Insights about Lactobacillar Sortase-Dependent Piliation
Abstract
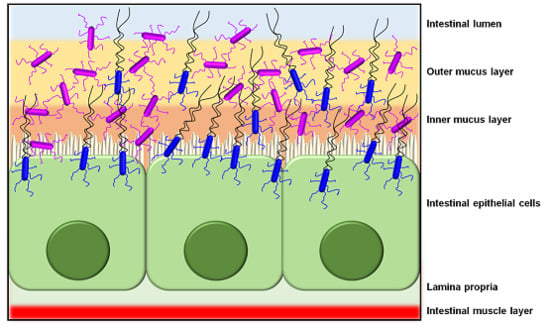
:1. Introduction
2. Sortase-Dependent Pilus: An Overview
2.1. Gene Expression
2.2. Assembly and Anchoring
2.3. Structural Composition
2.4. Functional Attributes
3. Sortase-Dependent Pili in Non-Pathogenic Lactobacilli
3.1. History of Discovery
3.2. Other Lactobacillar Pilus Operons
4. SpaCBA Piliation
4.1. Genetics
4.2. Structure
4.3. Adhesion
4.4. Immunogenicity
5. SpaFED Piliation
5.1. Genetics
5.2. Structure
5.3. Adhesion
5.4. Immunogenicity
6. LrpCBA Piliation
6.1. Genetics
6.2. Structure
6.3. Adhesion
6.4. Immunogenicity
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kline, K.A.; Dodson, K.W.; Caparon, M.G.; Hultgren, S.J. A tale of two pili: Assembly and function of pili in bacteria. Trends Microbiol. 2010, 18, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Proft, T.; Baker, E.N. Pili in Gram-negative and Gram-positive bacteria—Structure, assembly and their role in disease. Cell. Mol. Life Sci. 2009, 66, 613–635. [Google Scholar] [CrossRef] [PubMed]
- Duguid, J.P.; Smith, I.W.; Dempster, G.; Edmunds, P.N. Non-flagellar filamentous appendages (fimbriae) and haemagglutinating activity in Bacterium coli. J. Pathol. Bacteriol. 1955, 70, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Fronzes, R.; Remaut, H.; Waksman, G. Architectures and biogenesis of non-flagellar protein appendages in Gram-negative bacteria. EMBO J. 2008, 27, 2271–2280. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, R.; Otsuki, K. Some properties of the pili of Corynebacterium renale. J. Bacteriol. 1970, 101, 1063–1069. [Google Scholar] [PubMed]
- Yanagawa, R.; Otsuki, K.; Tokui, T. Electron microscopy of fine structure of Corynebacterium renale with special reference to pili. Jpn. J. Vet. Res. 1968, 16, 31–37. [Google Scholar] [PubMed]
- Baker, E.N.; Squire, C.J.; Young, P.G. Self-generated covalent cross-links in the cell-surface adhesins of Gram-positive bacteria. Biochem. Soc. Trans. 2015, 43, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Hilleringmann, M.; Ringler, P.; Müller, S.A.; de Angelis, G.; Rappuoli, R.; Ferlenghi, I.; Engel, A. Molecular architecture of Streptococcus pneumoniae TIGR4 pili. EMBO J. 2009, 28, 3921–3930. [Google Scholar] [CrossRef] [PubMed]
- Soriani, M.; Telford, J.L. Relevance of pili in pathogenic streptococci pathogenesis and vaccine development. Future Microbiol. 2010, 5, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Douillard, F.P.; Ribbera, A.; Järvinen, H.M.; Kant, R.; Pietilä, T.E.; Randazzo, C.; Paulin, L.; Laine, P.K.; Caggia, C.; von Ossowski, I.; et al. Comparative genomic and functional analysis of Lactobacillus casei and Lactobacillus rhamnosus strains marketed as probiotics. Appl. Environ. Microbiol. 2013, 79, 1923–1933. [Google Scholar] [CrossRef] [PubMed]
- Kankainen, M.; Paulin, L.; Tynkkynen, S.; von Ossowski, I.; Reunanen, J.; Partanen, P.; Satokari, R.; Vesterlund, S.; Hendrickx, A.P.; Lebeer, S.; et al. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc. Natl. Acad. Sci. USA 2009, 106, 17193–17198. [Google Scholar] [CrossRef] [PubMed]
- Kant, R.; Rintahaka, J.; Yu, X.; Sigvart-Mattila, P.; Paulin, L.; Mecklin, J.P.; Saarela, M.; Palva, A.; von Ossowski, I. A comparative pan-genome perspective of niche-adaptable cell-surface protein phenotypes in Lactobacillus rhamnosus. PLoS ONE 2014, 9, e102762. [Google Scholar] [CrossRef] [PubMed]
- Lebeer, S.; Verhoeven, T.L.; Francius, G.; Schoofs, G.; Lambrichts, I.; Dufrêne, Y.; Vanderleyden, J.; De Keersmaecker, S.C. Identification of a gene cluster for the biosynthesis of a long, galactose-rich exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase. Appl. Environ. Microbiol. 2009, 75, 3554–3563. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Jaatinen, A.; Rintahaka, J.; Hynönen, U.; Lyytinen, O.; Kant, R.; Åvall-Jääskeläinen, S.; von Ossowski, I.; Palva, A. Human gut-commensalic Lactobacillus ruminis ATCC 25644 displays sortase-assembled surface piliation: Phenotypic characterization of its fimbrial operon through in silico predictive analysis and recombinant expression in Lactococcus lactis. PLoS ONE 2015, 10, e0145718. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Serafini, F.; Foroni, E.; Duranti, S.; O’Connell Motherway, M.; Taverniti, V.; Mangifesta, M.; Milani, C.; Viappiani, A.; Roversi, T.; et al. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc. Natl. Acad. Sci. USA 2013, 110, 11151–11156. [Google Scholar] [CrossRef] [PubMed]
- Danne, C.; Dramsi, S. Pili of Gram-positive bacteria: Roles in host colonization. Res. Microbiol. 2012, 163, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Baker, E.N. Structure and assembly of Gram-positive bacterial pili: Unique covalent polymers. Curr. Opin. Struct. Biol. 2012, 22, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, V. Pilins in Gram-positive bacteria: A structural perspective. IUBMB Life 2015, 67, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Pansegrau, W.; Bagnoli, F. Pilus assembly in Gram-positive bacteria. Curr. Top. Microbiol. Immunol. 2017, 404, 203–233. [Google Scholar] [PubMed]
- Mandlik, A.; Swierczynski, A.; Das, A.; Ton-That, H. Pili in Gram-positive bacteria: Assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008, 16, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Budzik, J.M.; Marraffini, L.A.; Schneewind, O. Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol. Microbiol. 2007, 66, 495–510. [Google Scholar] [CrossRef] [PubMed]
- Danne, C.; Entenza, J.M.; Mallet, A.; Briandet, R.; Débarbouillé, M.; Nato, F.; Glaser, P.; Jouvion, G.; Moreillon, P.; Trieu-Cuot, P.; et al. Molecular characterization of a Streptococcus gallolyticus genomic island encoding a pilus involved in endocarditis. J. Infect. Dis. 2011, 204, 1960–1970. [Google Scholar] [CrossRef] [PubMed]
- Rosini, R.; Rinaudo, C.D.; Soriani, M.; Lauer, P.; Mora, M.; Maione, D.; Taddei, A.; Santi, I.; Ghezzo, C.; Brettoni, C.; et al. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol. Microbiol. 2006, 61, 126–141. [Google Scholar] [CrossRef] [PubMed]
- Ton-That, H.; Schneewind, O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 2003, 50, 1429–1438. [Google Scholar] [CrossRef] [PubMed]
- Bourgogne, A.; Singh, K.V.; Fox, K.A.; Pflughoeft, K.J.; Murray, B.E.; Garsin, D.A. EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF. J. Bacteriol. 2007, 189, 6490–6493. [Google Scholar] [CrossRef] [PubMed]
- Nallapareddy, S.R.; Singh, K.V.; Sillanpää, J.; Garsin, D.A.; Höök, M.; Erlandsen, S.L.; Murray, B.E. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Investig. 2006, 116, 2799–2807. [Google Scholar] [CrossRef] [PubMed]
- Nakata, M.; Köller, T.; Moritz, K.; Ribardo, D.; Jonas, L.; McIver, K.S.; Sumitomo, T.; Terao, Y.; Kawabata, S.; Podbielski, A.; et al. Mode of expression and functional characterization of FCT-3 pilus region-encoded proteins in Streptococcus pyogenes serotype M49. Infect. Immun. 2009, 77, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Barocchi, M.A.; Ries, J.; Zogaj, X.; Hemsley, C.; Albiger, B.; Kanth, A.; Dahlberg, S.; Fernebro, J.; Moschioni, M.; Masignani, V.; et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. USA 2006, 103, 2857–2862. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Ono, E.; Yanagawa, R. Comparison of surface hydrophobicity of piliated and non-piliated clones of Corynebacterium renale and Corynebacterium pilosum. Vet. Microbiol. 1987, 14, 165–171. [Google Scholar] [CrossRef]
- Basset, A.; Turner, K.H.; Boush, E.; Sayeed, S.; Dove, S.L.; Malley, R. Expression of the type 1 pneumococcal pilus is bistable and negatively regulated by the structural component RrgA. Infect. Immun. 2011, 79, 2974–2983. [Google Scholar] [CrossRef] [PubMed]
- Basset, A.; Turner, K.H.; Boush, E.; Sayeed, S.; Dove, S.L.; Malley, R. An epigenetic switch mediates bistable expression of the type 1 pilus genes in Streptococcus pneumoniae. J. Bacteriol. 2012, 194, 1088–1091. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, G.; Moschioni, M.; Muzzi, A.; Pezzicoli, A.; Censini, S.; Delany, I.; Lo Sapio, M.; Sinisi, A.; Donati, C.; Masignani, V.; et al. The Streptococcus pneumoniae pilus-1 displays a biphasic expression pattern. PLoS ONE 2011, 6, e21269. [Google Scholar] [CrossRef] [PubMed]
- Kreikemeyer, B.; Gámez, G.; Margarit, I.; Giard, J.C.; Hammerschmidt, S.; Hartke, A.; Podbielski, A. Genomic organization, structure, regulation and pathogenic role of pilus constituents in major pathogenic Streptococci and Enterococci. Int. J. Med. Microbiol. 2011, 301, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Hava, D.L.; Hemsley, C.J.; Camilli, A. Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA. J. Bacteriol. 2003, 185, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Danne, C.; Dubrac, S.; Trieu-Cuot, P.; Dramsi, S. Single cell stochastic regulation of pilus phase variation by an attenuation-like mechanism. PLoS Pathog. 2014, 10, e1003860. [Google Scholar] [CrossRef] [PubMed]
- Pancotto, L.; de Angelis, G.; Bizzarri, E.; Barocchi, M.A.; del Giudice, G.; Moschioni, M.; Ruggiero, P. Expression of the Streptococcus pneumoniae pilus-1 undergoes on and off switching during colonization in mice. Sci. Rep. 2013, 3, 2040. [Google Scholar] [CrossRef] [PubMed]
- Hendrickx, A.P.; Budzik, J.M.; Oh, S.Y.; Schneewind, O. Architects at the bacterial surface—Sortases and the assembly of pili with isopeptide bonds. Nat. Rev. Microbiol. 2011, 9, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Marraffini, L.A.; Dedent, A.C.; Schneewind, O. Sortases and the art of anchoring proteins to the envelopes of Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2006, 70, 192–221. [Google Scholar] [CrossRef] [PubMed]
- Spirig, T.; Weiner, E.M.; Clubb, R.T. Sortase enzymes in Gram-positive bacteria. Mol. Microbiol. 2011, 82, 1044–1059. [Google Scholar] [CrossRef] [PubMed]
- Schneewind, O.; Missiakas, D. Sec-secretion and sortase-mediated anchoring of proteins in Gram-positive bacteria. Biochim. Biophys. Acta 2014, 1843, 1687–1697. [Google Scholar] [CrossRef] [PubMed]
- Cozzi, R.; Malito, E.; Nuccitelli, A.; D’Onofrio, M.; Martinelli, M.; Ferlenghi, I.; Grandi, G.; Telford, J.L.; Maione, D.; Rinaudo, C.D. Structure analysis and site-directed mutagenesis of defined key residues and motives for pilus-related sortase C1 in group B Streptococcus. FASEB J. 2011, 25, 1874–1886. [Google Scholar] [CrossRef] [PubMed]
- Mazmanian, S.K.; Liu, G.; Ton-That, H.; Schneewind, O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 1999, 285, 760–763. [Google Scholar] [CrossRef] [PubMed]
- Khare, B.; Krishnan, V.; Rajashankar, K.R.; I-Hsiu, H.; Xin, M.; Ton-That, H.; Narayana, S.V. Structural differences between the Streptococcus agalactiae housekeeping and pilus-specific sortases: SrtA and SrtC1. PLoS ONE 2011, 6, e22995. [Google Scholar] [CrossRef] [PubMed]
- Necchi, F.; Nardi-Dei, V.; Biagini, M.; Assfalg, M.; Nuccitelli, A.; Cozzi, R.; Norais, N.; Telford, J.L.; Rinaudo, C.D.; Grandi, G.; et al. Sortase a substrate specificity in GBS pilus 2a cell wall anchoring. PLoS ONE 2011, 6, e25300. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, A.; Mandlik, A.; Swierczynski, A.; Gaspar, A.; Das, A.; Ton-That, H. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol. Microbiol. 2007, 66, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Budzik, J.M.; Oh, S.Y.; Schneewind, O. Cell wall anchor structure of BcpA pili in Bacillus anthracis. J. Biol. Chem. 2008, 283, 36676–36686. [Google Scholar] [CrossRef] [PubMed]
- Mandlik, A.; Das, A.; Ton-That, H. The molecular switch that activates the cell wall anchoring step of pilus assembly in Gram-positive bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 14147–14152. [Google Scholar] [CrossRef] [PubMed]
- Dramsi, S.; Caliot, E.; Bonne, I.; Guadagnini, S.; Prévost, M.C.; Kojadinovic, M.; Lalioui, L.; Poyart, C.; Trieu-Cuot, P. Assembly and role of pili in group B streptococci. Mol. Microbiol. 2006, 60, 1401–1413. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, A.H.; Ton-That, H. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J. Bacteriol. 2006, 188, 1526–1533. [Google Scholar] [CrossRef] [PubMed]
- Ton-That, H.; Marraffini, L.A.; Schneewind, O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol. Microbiol. 2004, 53, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Cozzi, R.; Nuccitelli, A.; D’Onofrio, M.; Necchi, F.; Rosini, R.; Zerbini, F.; Biagini, M.; Norais, N.; Beier, C.; Telford, J.L.; et al. New insights into the role of the glutamic acid of the E-box motif in group B Streptococcus pilus 2a assembly. FASEB J. 2012, 26, 2008–2018. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Baker, E.N. Intramolecular isopeptide bonds give thermodynamic and proteolytic stability to the major pilin protein of Streptococcus pyogenes. J. Biol. Chem. 2009, 284, 20729–20737. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Coulibaly, F.; Clow, F.; Proft, T.; Baker, E.N. Stabilizing isopeptide bonds revealed in Gram-positive bacterial pilus structure. Science 2007, 318, 1625–1628. [Google Scholar] [CrossRef] [PubMed]
- Deivanayagam, C.C.; Rich, R.L.; Carson, M.; Owens, R.T.; Danthuluri, S.; Bice, T.; Höök, M.; Narayana, S.V. Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure 2000, 8, 67–78. [Google Scholar] [CrossRef]
- Symersky, J.; Patti, J.M.; Carson, M.; House-Pompeo, K.; Teale, M.; Moore, D.; Jin, L.; Schneider, A.; DeLucas, L.J.; Höök, M.; et al. Structure of the collagen-binding domain from a Staphylococcus aureus adhesin. Nat. Struct. Biol. 1997, 4, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Paterson, N.G.; Gaspar, A.H.; Ton-That, H.; Baker, E.N. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc. Natl. Acad. Sci. USA 2009, 106, 16967–16971. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Baker, E.N. Intramolecular isopeptide bonds: Protein crosslinks built for stress? Trends Biochem. Sci. 2011, 36, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Izoré, T.; Contreras-Martel, C.; El Mortaji, L.; Manzano, C.; Terrasse, R.; Vernet, T.; Di Guilmi, A.M.; Dessen, A. Structural basis of host cell recognition by the pilus adhesin from Streptococcus pneumoniae. Structure 2010, 18, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, V.; Dwivedi, P.; Kim, B.J.; Samal, A.; Macon, K.; Ma, X.; Mishra, A.; Doran, K.S.; Ton-That, H.; Narayana, S.V. Structure of Streptococcus agalactiae tip pilin GBS104: A model for GBS pili assembly and host interactions. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1073–1089. [Google Scholar] [CrossRef] [PubMed]
- Linke-Winnebeck, C.; Paterson, N.G.; Young, P.G.; Middleditch, M.J.; Greenwood, D.R.; Witte, G.; Baker, E.N. Structural model for covalent adhesion of the Streptococcus pyogenes pilus through a thioester bond. J. Biol. Chem. 2014, 289, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Pointon, J.A.; Smith, W.D.; Saalbach, G.; Crow, A.; Kehoe, M.A.; Banfield, M.J. A highly unusual thioester bond in a pilus adhesin is required for efficient host cell interaction. J. Biol. Chem. 2010, 285, 33858–33866. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, V.; Gaspar, A.H.; Ye, N.; Mandlik, A.; Ton-That, H.; Narayana, S.V. An IgG-like domain in the minor pilin GBS52 of Streptococcus agalactiae mediates lung epithelial cell adhesion. Structure 2007, 15, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Linke, C.; Young, P.G.; Kang, H.J.; Bunker, R.D.; Middleditch, M.J.; Caradoc-Davies, T.T.; Proft, T.; Baker, E.N. Crystal structure of the minor pilin FctB reveals determinants of group A streptococcal pilus anchoring. J. Biol. Chem. 2010, 285, 20381–20389. [Google Scholar] [CrossRef] [PubMed]
- Shaik, M.M.; Maccagni, A.; Tourcier, G.; Di Guilmi, A.M.; Dessen, A. Structural basis of pilus anchoring by the ancillary pilin RrgC of Streptococcus pneumoniae. J. Biol. Chem. 2014, 289, 16988–16997. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Paterson, N.G.; Kim, C.U.; Middleditch, M.; Chang, C.; Ton-That, H.; Baker, E.N. A slow-forming isopeptide bond in the structure of the major pilin SpaD from Corynebacterium diphtheriae has implications for pilus assembly. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 1190–1201. [Google Scholar] [CrossRef] [PubMed]
- Vengadesan, K.; Narayana, S.V. Structural biology of Gram-positive bacterial adhesins. Protein Sci. 2011, 20, 759–772. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H.V.; Guiton, P.S.; Kline, K.A.; Port, G.C.; Pinkner, J.S.; Neiers, F.; Normark, S.; Henriques-Normark, B.; Caparon, M.G.; Hultgren, S.J. The metal ion-dependent adhesion site motif of the Enterococcus faecalis EbpA pilin mediates pilus function in catheter-associated urinary tract infection. MBio 2012, 3, e00177-12. [Google Scholar] [CrossRef] [PubMed]
- Konto-Ghiorghi, Y.; Mairey, E.; Mallet, A.; Duménil, G.; Caliot, E.; Trieu-Cuot, P.; Dramsi, S. Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 2009, 5, e1000422. [Google Scholar] [CrossRef] [PubMed]
- Becherelli, M.; Manetti, A.G.; Buccato, S.; Viciani, E.; Ciucchi, L.; Mollica, G.; Grandi, G.; Margarit, I. The ancillary protein 1 of Streptococcus pyogenes FCT-1 pili mediates cell adhesion and biofilm formation through heterophilic as well as homophilic interactions. Mol. Microbiol. 2012, 83, 1035–1047. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, C.A.; Hynes, R.O. Distribution and evolution of von Willebrand/integrin a domains: Widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 2002, 13, 3369–3387. [Google Scholar] [CrossRef] [PubMed]
- Hilleringmann, M.; Giusti, F.; Baudner, B.C.; Masignani, V.; Covacci, A.; Rappuoli, R.; Barocchi, M.A.; Ferlenghi, I. Pneumococcal pili are composed of protofilaments exposing adhesive clusters of RrgA. PLoS Pathog. 2008, 4, e1000026. [Google Scholar] [CrossRef] [PubMed]
- Nallapareddy, S.R.; Sillanpää, J.; Mitchell, J.; Singh, K.V.; Chowdhury, S.A.; Weinstock, G.M.; Sullam, P.M.; Murray, B.E. Conservation of Ebp-type pilus genes among Enterococci and demonstration of their role in adherence of Enterococcus faecalis to human platelets. Infect. Immun. 2011, 79, 2911–2920. [Google Scholar] [CrossRef] [PubMed]
- Sillanpää, J.; Nallapareddy, S.R.; Qin, X.; Singh, K.V.; Muzny, D.M.; Kovar, C.L.; Nazareth, L.V.; Gibbs, R.A.; Ferraro, M.J.; Steckelberg, J.M.; et al. A collagen-binding adhesin, Acb, and ten other putative MSCRAMM and pilus family proteins of Streptococcus gallolyticus subsp. gallolyticus (Streptococcus bovis group, biotype I). J. Bacteriol. 2009, 191, 6643–6653. [Google Scholar]
- Abbot, E.L.; Smith, W.D.; Siou, G.P.; Chiriboga, C.; Smith, R.J.; Wilson, J.A.; Hirst, B.H.; Kehoe, M.A. Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cell. Microbiol. 2007, 9, 1822–1833. [Google Scholar] [CrossRef] [PubMed]
- Mandlik, A.; Swierczynski, A.; Das, A.; Ton-That, H. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol. Microbiol. 2007, 64, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Manetti, A.G.; Zingaretti, C.; Falugi, F.; Capo, S.; Bombaci, M.; Bagnoli, F.; Gambellini, G.; Bensi, G.; Mora, M.; Edwards, A.M.; et al. Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol. Microbiol. 2007, 64, 968–983. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; Aymeric, L.; du Merle, L.; Danne, C.; Robbe-Masselot, C.; Trieu-Cuot, P.; Sansonetti, P.; Dramsi, S. Streptococcus gallolyticus Pil3 pilus is required for adhesion to colonic mucus and for colonization of mouse distal colon. J. Infect. Dis. 2015, 212, 1646–1655. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; Porrini, C.; du Merle, L.; Danne, C.; Robbe-Masselot, C.; Trieu-Cuot, P.; Dramsi, S. The pil3 pilus of Streptococcus gallolyticus binds to intestinal mucins and to fibrinogen. Gut Microbes 2016, 7, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Devarajan, B.; Reardon, M.E.; Dwivedi, P.; Krishnan, V.; Cisar, J.O.; Das, A.; Narayana, S.V.; Ton-That, H. Two autonomous structural modules in the fimbrial shaft adhesin FimA mediate Actinomyces interactions with streptococci and host cells during oral biofilm development. Mol. Microbiol. 2011, 81, 1205–1220. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Wu, C.; Yang, J.; Cisar, J.O.; Das, A.; Ton-That, H. The Actinomyces oris type 2 fimbrial shaft FimA mediates co-aggregation with oral streptococci, adherence to red blood cells and biofilm development. Mol. Microbiol. 2010, 77, 841–854. [Google Scholar] [CrossRef] [PubMed]
- Okahashi, N.; Nakata, M.; Terao, Y.; Isoda, R.; Sakurai, A.; Sumitomo, T.; Yamaguchi, M.; Kimura, R.K.; Oiki, E.; Kawabata, S.; et al. Pili of oral Streptococcus sanguinis bind to salivary amylase and promote the biofilm formation. Microb. Pathog. 2011, 50, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, D.G.; Bowler, P.G. Biofilm delays wound healing: A review of the evidence. Burns Trauma 2013, 1, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Basset, A.; Zhang, F.; Benes, C.; Sayeed, S.; Herd, M.; Thompson, C.; Golenbock, D.T.; Camilli, A.; Malley, R. Toll-like receptor (TLR) 2 mediates inflammatory responses to oligomerized RrgA pneumococcal pilus type 1 protein. J. Biol. Chem. 2013, 288, 2665–2675. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, D.; Carey, A.J.; Caliot, E.; Webb, R.I.; Layton, J.R.; Wang, Y.; Bohnsack, J.F.; Adderson, E.E.; Ulett, G.C. Phylogenetic lineage and pilus protein Spb1/SAN1518 affect opsonin-independent phagocytosis and intracellular survival of group B Streptococcus. Microbes Infect. 2011, 13, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Maisey, H.C.; Quach, D.; Hensler, M.E.; Liu, G.Y.; Gallo, R.L.; Nizet, V.; Doran, K.S. A group B streptococcal pilus protein promotes phagocyte resistance and systemic virulence. FASEB J. 2008, 22, 1715–1724. [Google Scholar] [CrossRef] [PubMed]
- Papasergi, S.; Brega, S.; Mistou, M.Y.; Firon, A.; Oxaran, V.; Dover, R.; Teti, G.; Shai, Y.; Trieu-Cuot, P.; Dramsi, S. The GBS PI-2a pilus is required for virulence in mice neonates. PLoS ONE 2011, 6, e18747. [Google Scholar] [CrossRef] [PubMed]
- Pace, F.; Pace, M.; Quartarone, G. Probiotics in digestive diseases: Focus on Lactobacillus GG. Minerva Gastroenterol. Dietol. 2015, 61, 273–292. [Google Scholar] [PubMed]
- Ton-That, H.; Schneewind, O. Assembly of pili in Gram-positive bacteria. Trends Microbiol. 2004, 12, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Huang, I.H.; Hendrickx, A.P.; Ton-That, H. Visualization of Gram-positive bacterial pili. Methods Mol. Biol. 2013, 966, 77–95. [Google Scholar] [PubMed]
- Reunanen, J.; von Ossowski, I.; Hendrickx, A.P.; Palva, A.; de Vos, W.M. Characterization of the SpaCBA pilus fibers in the probiotic Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 2012, 78, 2337–2344. [Google Scholar] [CrossRef] [PubMed]
- Walker, A. Genome watch: Probiotics stick it to the man. Nat. Rev. Microbiol. 2009, 7, 843. [Google Scholar] [CrossRef] [PubMed]
- Broadbent, J.R.; Neeno-Eckwall, E.C.; Stahl, B.; Tandee, K.; Cai, H.; Morovic, W.; Horvath, P.; Heidenreich, J.; Perna, N.T.; Barrangou, R.; et al. Analysis of the Lactobacillus casei supragenome and its influence in species evolution and lifestyle adaptation. BMC Genom. 2012, 13, 533. [Google Scholar] [CrossRef] [PubMed]
- Smokvina, T.; Wels, M.; Polka, J.; Chervaux, C.; Brisse, S.; Boekhorst, J.; van Hylckama Vlieg, J.E.; Siezen, R.J. Lactobacillus paracasei comparative genomics: Towards species pan-genome definition and exploitation of diversity. PLoS ONE 2013, 8, e68731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toh, H.; Oshima, K.; Nakano, A.; Takahata, M.; Murakami, M.; Takaki, T.; Nishiyama, H.; Igimi, S.; Hattori, M.; Morita, H. Genomic adaptation of the Lactobacillus casei group. PLoS ONE 2013, 8, e75073. [Google Scholar] [CrossRef] [PubMed]
- Forde, B.M.; Neville, B.A.; O’Donnell, M.M.; Riboulet-Bisson, E.; Claesson, M.J.; Coghlan, A.; Ross, R.P.; O’Toole, P.W. Genome sequences and comparative genomics of two Lactobacillus ruminis strains from the bovine and human intestinal tracts. Microb. Cell. Fact. 2011, 10, S13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Z.; Harris, H.M.; McCann, A.; Guo, C.; Argimón, S.; Zhang, W.; Yang, X.; Jeffery, I.B.; Cooney, J.C.; Kagawa, T.F.; et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 2015, 6, 8322. [Google Scholar] [CrossRef] [PubMed]
- Douillard, F.P.; Ribbera, A.; Kant, R.; Pietilä, T.E.; Järvinen, H.M.; Messing, M.; Randazzo, C.L.; Paulin, L.; Laine, P.; Ritari, J.; et al. Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG. PLoS Genet. 2013, 9, e1003683. [Google Scholar] [CrossRef] [PubMed]
- Rintahaka, J.; Yu, X.; Kant, R.; Palva, A.; von Ossowski, I. Phenotypical analysis of the Lactobacillus rhamnosus GG fimbrial spaFED operon: Surface expression and functional characterization of recombinant SpaFED pili in Lactococcus lactis. PLoS ONE 2014, 9, e113922. [Google Scholar] [CrossRef] [PubMed]
- Douillard, F.P.; Ribbera, A.; Xiao, K.; Ritari, J.; Rasinkangas, P.; Paulin, L.; Palva, A.; Hao, Y.; de Vos, W.M. Polymorphisms, chromosomal rearrangements, and mutator phenotype development during experimental evolution of Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 2016, 82, 3783–3792. [Google Scholar] [CrossRef] [PubMed]
- Chaurasia, P.; Pratap, S.; von Ossowski, I.; Palva, A.; Krishnan, V. New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit. Sci. Rep. 2016, 6, 28664. [Google Scholar] [CrossRef] [PubMed]
- Douillard, F.P.; Rasinkangas, P.; von Ossowski, I.; Reunanen, J.; Palva, A.; de Vos, W.M. Functional identification of conserved residues involved in Lactobacillus rhamnosus strain GG sortase specificity and pilus biogenesis. J. Biol. Chem. 2014, 289, 15764–15775. [Google Scholar] [CrossRef] [PubMed]
- Cozzi, R.; Prigozhin, D.; Rosini, R.; Abate, F.; Bottomley, M.J.; Grandi, G.; Telford, J.L.; Rinaudo, C.D.; Maione, D.; Alber, T. Structural basis for group B Streptococcus pilus 1 sortases C regulation and specificity. PLoS ONE 2012, 7, e49048. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; von Ossowski, I.; Palva, A.; Krishnan, V. Purification, crystallization and preliminary crystallographic analysis of the SpaA backbone-pilin subunit from probiotic Lactobacillus rhamnosus GG. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 1182–1185. [Google Scholar] [CrossRef] [PubMed]
- Von Ossowski, I.; Pietilä, T.E.; Rintahaka, J.; Nummenmaa, E.; Mäkinen, V.M.; Reunanen, J.; Satokari, R.; de Vos, W.M.; Palva, I.; Palva, A. Using recombinant lactococci as an approach to dissect the immunomodulating capacity of surface piliation in probiotic Lactobacillus rhamnosus GG. PLoS ONE 2013, 8, e64416. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, K.; Ueno, S.; Sugiyama, M.; Yamamoto, Y.; Mukai, T. Lactobacillus rhamnosus GG SpaC pilin subunit binds to the carbohydrate moieties of intestinal glycoconjugates. Anim. Sci. J. 2016, 87, 809–815. [Google Scholar] [CrossRef] [PubMed]
- Lebeer, S.; Claes, I.; Tytgat, H.L.; Verhoeven, T.L.; Marien, E.; von Ossowski, I.; Reunanen, J.; Palva, A.; Vos, W.M.; Keersmaecker, S.C.; et al. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 2012, 78, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Ardita, C.S.; Mercante, J.W.; Kwon, Y.M.; Luo, L.; Crawford, M.E.; Powell, D.N.; Jones, R.M.; Neish, A.S. Epithelial adhesion mediated by pilin SpaC is required for Lactobacillus rhamnosus GG-induced cellular responses. Appl. Environ. Microbiol. 2014, 80, 5068–5077. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Beaussart, A.; Alsteens, D.; Dupres, V.; Claes, I.; von Ossowski, I.; de Vos, W.M.; Palva, A.; Lebeer, S.; Vanderleyden, J.; et al. Adhesion and nanomechanics of pili from the probiotic Lactobacillus rhamnosus GG. ACS Nano 2013, 7, 3685–3697. [Google Scholar] [CrossRef] [PubMed]
- Kant, A.; von Ossowski, I.; Palva, A.; Krishnan, V. Crystallization and X-ray crystallographic analysis of the adhesive SpaC pilin subunit in the SpaCBA pilus of gut-adapted Lactobacillus rhamnosus GG. Protein Pept. Lett. 2016, 23, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Dupres, V.; Beaussart, A.; Lebeer, S.; Claes, I.J.; Vanderleyden, J.; Dufrêne, Y.F. Deciphering the nanometer-scale organization and assembly of Lactobacillus rhamnosus GG pili using atomic force microscopy. Langmuir 2012, 28, 2211–2216. [Google Scholar] [CrossRef] [PubMed]
- Sullan, R.M.; Beaussart, A.; Tripathi, P.; Derclaye, S.; El-Kirat-Chatel, S.; Li, J.K.; Schneider, Y.J.; Vanderleyden, J.; Lebeer, S.; Dufrêne, Y.F. Single-cell force spectroscopy of pili-mediated adhesion. Nanoscale 2014, 6, 1134–1143. [Google Scholar] [CrossRef] [PubMed]
- Von Ossowski, I.; Reunanen, J.; Satokari, R.; Vesterlund, S.; Kankainen, M.; Huhtinen, H.; Tynkkynen, S.; Salminen, S.; de Vos, W.M.; Palva, A. Mucosal adhesion properties of the probiotic Lactobacillus rhamnosus GG SpaCBA and SpaFED pilin subunits. Appl. Environ. Microbiol. 2010, 76, 2049–2057. [Google Scholar] [CrossRef] [PubMed]
- Tytgat, H.L.; Douillard, F.P.; Reunanen, J.; Rasinkangas, P.; Hendrickx, A.P.; Laine, P.K.; Paulin, L.; Satokari, R.; de Vos, W.M. Lactobacillus rhamnosus GG outcompetes Enterococcus faecium via mucus-binding pili: Evidence for a novel and heterospecific probiotic mechanism. Appl. Environ. Microbiol. 2016, 82, 5756–5762. [Google Scholar] [CrossRef] [PubMed]
- Ganguli, K.; Collado, M.C.; Rautava, J.; Lu, L.; Satokari, R.; von Ossowski, I.; Reunanen, J.; de Vos, W.M.; Palva, A.; Isolauri, E.; et al. Lactobacillus rhamnosus GG and its SpaC pilus adhesin modulate inflammatory responsiveness and TLR-related gene expression in the fetal human gut. Pediatr. Res. 2015, 77, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Vargas García, C.E.; Petrova, M.; Claes, I.J.; De Boeck, I.; Verhoeven, T.L.; Dilissen, E.; von Ossowski, I.; Palva, A.; Bullens, D.M.; Vanderleyden, J.; et al. Piliation of Lactobacillus rhamnosus GG promotes adhesion, phagocytosis, and cytokine modulation in macrophages. Appl. Environ. Microbiol. 2015, 81, 2050–2062. [Google Scholar] [CrossRef] [PubMed]
- Tytgat, H.L.; van Teijlingen, N.H.; Sullan, R.M.; Douillard, F.P.; Rasinkangas, P.; Messing, M.; Reunanen, J.; Satokari, R.; Vanderleyden, J.; Dufrêne, Y.F.; et al. Probiotic gut microbiota isolate interacts with dendritic cells via glycosylated heterotrimeric pili. PLoS ONE 2016, 11, e0151824. [Google Scholar] [CrossRef] [PubMed]
- Cheesman, S.E.; Guillemin, K. We know you are in there: Conversing with the indigenous gut microbiota. Res. Microbiol. 2007, 158, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Rakoff-Nahoum, S.; Medzhitov, R. Role of the innate immune system and host-commensal mutualism. Curr. Top. Microbiol. Immunol. 2006, 308, 1–18. [Google Scholar] [PubMed]
- Rakoff-Nahoum, S.; Paglino, J.; Eslami-Varzaneh, F.; Edberg, S.; Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004, 118, 229–241. [Google Scholar] [CrossRef] [PubMed]
- O’Connell Motherway, M.; Zomer, A.; Leahy, S.C.; Reunanen, J.; Bottacini, F.; Claesson, M.J.; O’Brien, F.; Flynn, K.; Casey, P.G.; Munoz, J.A.; et al. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. USA 2011, 108, 11217–11222. [Google Scholar] [CrossRef] [PubMed]
- Chaurasia, P.; von Ossowski, I.; Palva, A.; Krishnan, V. Purification, crystallization and preliminary X-ray diffraction analysis of SpaD, a backbone-pilin subunit encoded by the fimbrial spaFED operon in Lactobacillus rhamnosus GG. Acta Crystallogr. F Struct. Biol. Commun. 2015, 71, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Megta, A.K.; Palva, A.; von Ossowski, I.; Krishnan, V. Crystallization and X-ray diffraction analysis of SpaE, a basal pilus protein from the gut-adapted Lactobacillus rhamnosus GG. Acta Crystallogr. F Struct. Biol. Commun. 2017, 73, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Neville, B.A.; Forde, B.M.; Claesson, M.J.; Darby, T.; Coghlan, A.; Nally, K.; Ross, R.P.; O’Toole, P.W. Characterization of pro-inflammatory flagellin proteins produced by Lactobacillus ruminis and related motile lactobacilli. PLoS ONE 2012, 7, e40592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Donnell, M.M.; Harris, H.M.; Lynch, D.B.; Ross, R.P.; O’Toole, P.W. Lactobacillus ruminis strains cluster according to their mammalian gut source. BMC Microbiol. 2015, 15, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvetti, E.; Torriani, S.; Felis, G.E. The genus Lactobacillus: A taxonomic update. Probiotics Antimicrob. Proteins 2012, 4, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, M.E.; Latham, M.J.; Garvie, E.I.; Zirngibl, J.; Kandler, O. Two new species of Lactobacillus isolated from the bovine rumen, Lactobacillus ruminis sp.nov. And Lactobacillus vitulinus sp.nov. J. Gen. Microbiol. 1973, 77, 37–49. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, J.; O’Toole, P.W. Lactobacillus: Host-microbe relationships. Curr. Top. Microbiol. Immunol. 2013, 358, 119–154. [Google Scholar] [PubMed]
- Reuter, G. The Lactobacillus and Bifidobacterium microflora of the human intestine: Composition and succession. Curr. Issues Intest. Microbiol. 2001, 2, 43–53. [Google Scholar] [PubMed]
- Kant, R.; Palva, A.; von Ossowski, I. An in silico pan-genomic probe for the molecular traits behind Lactobacillus ruminis gut autochthony. PLoS ONE 2017, 12, e0175541. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Åvall-Jääskeläinen, S.; Koort, J.; Lindholm, A.; Rintahaka, J.; von Ossowski, I.; Palva, A.; Hynönen, U. A comparative characterization of different host-sourced Lactobacillus ruminis strains and their adhesive, inhibitory, and immunomodulating functions. Front. Microbiol. 2017, 8, 657. [Google Scholar] [CrossRef] [PubMed]
- Manetti, A.G.O.; Spadafina, T. The role of pili in the formation of biofilm and bacterial communities. In Bacterial Pili: Structure, Synthesis and Role in Disease, 1st ed.; Barocchi, M.A., Telford, J.L., Eds.; CAB International: Wallingford, UK, 2013; Volume 27, pp. 151–164. [Google Scholar]
- Rosini, R.; Margarit, I. Biofilm formation by Streptococcus agalactiae: Influence of environmental conditions and implicated virulence factors. Front. Cell. Infect. Microbiol. 2015, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Taweechotipatr, M.; Iyer, C.; Spinler, J.K.; Versalovic, J.; Tumwasorn, S. Lactobacillus saerimneri and Lactobacillus ruminis: Novel human-derived probiotic strains with immunomodulatory activities. FEMS Microbiol. Lett. 2009, 293, 65–72. [Google Scholar] [CrossRef] [PubMed]




| Primary Structure Feature | Tip Pilin Subunit |
|---|---|
| N-terminal secretory domain | MTAKVARTGHLFAVLLILMSMLTGLVTSGSSVVT (SpaC) LPRKWIHMLMLLLMLVTQIGSA (SpaF) MERNKIFKKLLCILGAVATVFAIVFAMGKFDGEKANA (LrpC) |
| C-terminal LPXTG domain | LPHTGGQGYQRLLGIALGLISAAFLLLLVVLIKRRVVKQHD (SpaC) LPKTGGSGILLFLMVAISACGGGWLLYLYLKRKEAR (SpaF) LPQTGGPGRLLFEALGSLLIVVACALTEVLIWRRIRSSKGV (LrpC) |
| Pilin motif | not detected (SpaC) not detected (SpaF) not detected (LrpC) |
| E-box | YGIQEAAAPTGY and YTMSETKAPDGY (SpaC) YRLTETKAPAGF (SpaF) YKLVETRTQSGY (LrpC) |
| Primary Structure Feature | Basal Pilin Subunit |
| N-terminal secretory domain | MTKSFRPLVILTFCLALLVSLATTTLQQTQA (SpaB) MRRFYWWLVPLLLLIGIVLGNTPHWVHA (SpaE) MKRVLKLLFMIVAFMTAVFAGSGQASA (LrpB) |
| C-terminal LPXTG domain | LPQTGDTVAAWLSVLGLIIFATVLAFNIKNQKINKWER (SpaB) LPAMSDWRNLRFVLLGSLLLLLATYFFIKNKKARHHACK (SpaE) LPQTGEAKSIMALLGIGIICLVVLVSVGRRNYKEEH (LrpB) |
| Pilin motif | TADFWQLVSKN (SpaB) PLQTIHLYPKN (SpaE) FPLGGQSYAKN (LrpB) |
| E-box | YLFKETAAPKNI (SpaB) YFFEELQGVPGY (SpaE) YYFSEVQAPKGY (LrpB) |
| Primary Structure Feature | Backbone Pilin Subunit |
| N-terminal secretory domain | MKKTIAKKVLTLTSTILMTLLMVLGFNGTRVQA (SpaA) MQVTFKKIGHSLLAALMLMSFLLPLLSAGKPVHA (SpaD) MKNHKKLRNALATLLLALPLALQGAVGVKTAQA (LrpA) |
| C-terminal LPXTG domain | LPHTGGTGTVIFAILGVALIAFGAVAYRKRRNGF (SpaA) LPMTGGIGLFAFLMIGAILMGGGHLMKKKTSKKV (SpaD) LPSTGGMGIVLFIAAGVVVMAGAAGTMIVRRNRRENI (LrpA) |
| Pilin motif | ADGNVYVYPKN (SpaA) DLTNIHLYPKD (SpaD) VQKSINIYPKN (LrpA) |
| E-box | YLFHETNPRAGY and YTAVETNVPDGY (SpaA) YAFHEAVTPQPY and YTLVETAAPEGY (SpaD) YLFAETDAPANI and YAVKEVKAPTGY (LrpA) |
© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Von Ossowski, I. Novel Molecular Insights about Lactobacillar Sortase-Dependent Piliation. Int. J. Mol. Sci. 2017, 18, 1551. https://doi.org/10.3390/ijms18071551
Von Ossowski I. Novel Molecular Insights about Lactobacillar Sortase-Dependent Piliation. International Journal of Molecular Sciences. 2017; 18(7):1551. https://doi.org/10.3390/ijms18071551
Chicago/Turabian StyleVon Ossowski, Ingemar. 2017. "Novel Molecular Insights about Lactobacillar Sortase-Dependent Piliation" International Journal of Molecular Sciences 18, no. 7: 1551. https://doi.org/10.3390/ijms18071551
APA StyleVon Ossowski, I. (2017). Novel Molecular Insights about Lactobacillar Sortase-Dependent Piliation. International Journal of Molecular Sciences, 18(7), 1551. https://doi.org/10.3390/ijms18071551