DNA Damage Tolerance by Eukaryotic DNA Polymerase and Primase PrimPol
Abstract
:1. Introduction
2. Activities and Fidelity of PrimPol
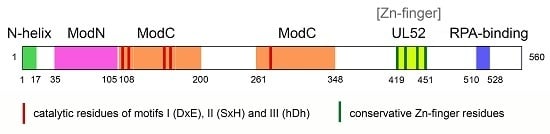
3. Structure of PrimPol
4. Functions of PrimPol in Cells
4.1. The Role of PrimPol in Nuclear Replication and DNA Translesion Synthesis
4.2. Functions of PrimPol in Mitochondria
5. Regulation of PrimPol Activity in Cells
6. PrimPol Dysfunction and Disease
7. Conclusions
Acknowledgments
Conflicts of Interest
References
- Lujan, S.A.; Williams, J.S.; Kunkel, T.A. DNA Polymerases Divide the Labor of Genome Replication. Trends Cell Biol. 2016, 26, 640–654. [Google Scholar] [CrossRef] [PubMed]
- Baranovskiy, A.G.; Tahirov, T.H. Elaborated Action of the Human Primosome. Genes 2017, 8, 62. [Google Scholar] [CrossRef] [PubMed]
- Harrington, C.; Perrino, F.W. Initiation of RNA-primed DNA synthesis in vitro by DNA polymerase α-primase. Nucleic Acids Res. 1995, 23, 1003–1009. [Google Scholar] [CrossRef] [PubMed]
- Ropp, P.A.; Copeland, W.C. Cloning and characterization of the human mitochondrial DNA polymerase, DNA polymerase γ. Genomics 1996, 36, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Wanrooij, S.; Falkenberg, M. The human mitochondrial replication fork in health and disease. Biochim. Biophys. Acta 2010, 1797, 1378–1388. [Google Scholar] [CrossRef] [PubMed]
- Young, M.J.; Copeland, W.C. Human mitochondrial DNA replication machinery and disease. Curr. Opin. Genet. Dev. 2016, 38, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Cadet, J.; Wagner, J.R. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb. Perspect. Biol. 2013, 5, a012559. [Google Scholar] [CrossRef] [PubMed]
- Ignatov, A.V.; Bondarenko, K.A.; Makarova, A.V. Non-bulky lesions in human DNA: Ways of formations, repair and replication. Acta Nat. 2017, in press. [Google Scholar]
- Irigaray, P.; Belpomme, D. Basic properties and molecular mechanisms of exogenous chemical carcinogens. Carcinogenesis 2010, 31, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Mao, P.; Wyrick, J.J.; Roberts, S.A.; Smerdon, M.J. UV-Induced DNA Damage and Mutagenesis in Chromatin. Photochem. Photobiol. 2017, 93, 216–228. [Google Scholar] [CrossRef] [PubMed]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Makarova, A.V.; Burgers, P.M. Eukaryotic DNA polymerase ζ. DNA Repair 2015, 29, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Vaisman, A.; Woodgate, R. Translesion DNA polymerases in eukaryotes: What makes them tick? Crit. Rev. Biochem. Mol. Biol. 2017, 52, 274–303. [Google Scholar] [CrossRef] [PubMed]
- Beard, W.A.; Wilson, S.H. Structures of human DNA polymerases ν and θ expose their end game. Nat. Struct. Mol. Biol. 2015, 22, 273–275. [Google Scholar] [CrossRef] [PubMed]
- Belousova, E.A.; Lavrik, O.I. DNA polymerases β and λ and their roles in cell. DNA Repair 2015, 29, 112–126. [Google Scholar] [CrossRef] [PubMed]
- Yamtich, J.; Sweasy, J.B. DNA polymerase family X: Function, structure, and cellular roles. Biochim. Biophys. Acta 2010, 1804, 1136–1150. [Google Scholar] [CrossRef] [PubMed]
- Yousefzadeh, M.J.; Wood, R.D. DNA polymerase POLQ and cellular defense against DNA damage. DNA Repair 2013, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Boyer, A.S.; Grgurevic, S.; Cazaux, C.; Hoffmann, J.S. The human specialized DNA polymerases and non-B DNA: Vital relationships to preserve genome integrity. J. Mol. Biol. 2013, 425, 4767–4781. [Google Scholar] [CrossRef] [PubMed]
- Helmrich, A.; Ballarino, M.; Nudler, E.; Tora, L. Transcription-replication encounters, consequences and genomic instability. Nat. Struct. Mol. Biol. 2013, 20, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Vasquez, K.M. Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability. DNA Repair 2014, 19, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Baranovskiy, A.G.; Babayeva, N.D.; Zhang, Y.; Gu, J.; Suwa, Y.; Pavlov, Y.I.; Tahirov, T.H. Mechanism of Concerted RNA-DNA Primer Synthesis by the Human Primosome. J. Biol. Chem. 2016, 291, 10006–10020. [Google Scholar] [CrossRef] [PubMed]
- Baranovskiy, A.G.; Zhang, Y.; Suwa, Y.; Babayeva, N.D.; Gu, J.; Pavlov, Y.I.; Tahirov, T.H. Crystal structure of the human primase. J. Biol. Chem. 2015, 290, 5635–5646. [Google Scholar] [CrossRef] [PubMed]
- Kilkenny, M.L.; Longo, M.A.; Perera, R.L.; Pellegrini, L. Structures of human primase reveal design of nucleotide elongation site and mode of Pol α tethering. Proc. Natl. Acad. Sci. USA 2013, 110, 15961–15966. [Google Scholar] [CrossRef] [PubMed]
- Guilliam, T.A.; Keen, B.A.; Brissett, N.C.; Doherty, A.J. Primase-polymerases are a functionally diverse superfamily of replication and repair enzymes. Nucleic Acids Res. 2015, 43, 6651–6664. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Koonin, E.V.; Leipe, D.D.; Aravind, L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: Structural insights and new members. Nucleic Acids Res. 2005, 33, 3875–3896. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, J.; Rudd, S.G.; Jozwiakowski, S.K.; Bailey, L.J.; Soura, V.; Taylor, E.; Stevanovic, I.; Green, A.J.; Stracker, T.H.; Lindsay, H.D.; et al. PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol. Cell 2013, 52, 566–573. [Google Scholar] [CrossRef] [PubMed]
- García-Gómez, S.; Reyes, A.; Martínez-Jiménez, M.I.; Chocrón, E.S.; Mourón, S.; Terrados, G.; Powell, C.; Salido, E.; Méndez, J.; Holt, I.J.; et al. PrimPol, an archaic primase/polymerase operating in human cells. Mol. Cell 2013, 52, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Lou, J.; Xia, Y.; Su, B.; Liu, T.; Cui, J.; Sun, Y.; Lou, H.; Huang, J. hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity. EMBO Rep. 2013, 14, 1104–11012. [Google Scholar] [CrossRef] [PubMed]
- Rudd, S.G.; Glover, L.; Jozwiakowski, S.K.; Horn, D.; Doherty, A.J. PPL2 translesion polymerase is essential for the completion of chromosomal DNA replication in the African trypanosome. Mol. Cell 2013, 52, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Guilliam, T.A.; Jozwiakowski, S.K.; Ehlinger, A.; Barnes, R.P.; Rudd, S.G.; Bailey, L.J.; Skehel, J.M.; Eckert, K.A.; Chazin, W.J.; Doherty, A.J. Human PrimPol is a highly error-prone polymerase regulated by single-stranded DNA binding proteins. Nucleic Acids Res. 2015, 43, 1056–1068. [Google Scholar] [CrossRef] [PubMed]
- Mislak, A.C.; Anderson, K.S. Insights into the Molecular Mechanism of Polymerization and Nucleoside Reverse Transcriptase Inhibitor Incorporation by Human PrimPol. Antimicrob. Agents Chemother. 2015, 60, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Tokarsky, E.J.; Wallenmeyer, P.C.; Phi, K.K.; Suo, Z. Significant impact of divalent metal ions on the fidelity, sugar selectivity, and drug incorporation efficiency of human PrimPol. DNA Repair 2017, 49, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Zafar, M.K.; Ketkar, A.; Lodeiro, M.F.; Cameron, C.E.; Eoff, R.L. Kinetic analysis of human PrimPol DNA polymerase activity reveals a generally error-prone enzyme capable of accurately bypassing 7,8-dihydro-8-oxo-2′-deoxyguanosine. Biochemistry 2014, 53, 6584–6594. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Jiménez, M.I.; García-Gómez, S.; Bebenek, K.; Sastre-Moreno, G.; Calvo, P.A.; Díaz-Talavera, A.; Kunkel, T.A.; Blanco, L. Alternative solutions and new scenarios for translesion DNA synthesis by human PrimPol. DNA Repair 2015, 29, 127–138. [Google Scholar] [CrossRef]
- Keen, B.A.; Jozwiakowski, S.K.; Bailey, L.J.; Bianchi, J.; Doherty, A.J. Molecular dissection of the domain architecture and catalytic activities of human PrimPol. Nucleic Acids Res. 2014, 42, 5830–5845. [Google Scholar] [CrossRef] [PubMed]
- Rechkoblit, O.; Gupta, Y.K.; Malik, R.; Rajashankar, K.R.; Johnson, R.E.; Prakash, L.; Prakash, S.; Aggarwal, A.K. Structure and mechanism of human PrimPol, a DNA polymerase with primase activity. Sci. Adv. 2016, 2, e1601317. [Google Scholar] [CrossRef] [PubMed]
- Frank, E.G.; Woodgate, R. Increased catalytic activity and altered fidelity of human DNA polymerase iota in the presence of manganese. J. Biol. Chem. 2007, 282, 24689–24696. [Google Scholar] [CrossRef] [PubMed]
- Makarova, A.V.; Ignatov, A.; Miropolskaya, N.; Kulbachinskiy, A. Roles of the active site residues and metal cofactors in noncanonical base-pairing during catalysis by human DNA polymerase iota. DNA Repair 2014, 22, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Blanca, G.; Shevelev, I.; Ramadan, K.; Villani, G.; Spadari, S.; Hubscher, U.; Maga, G. Human DNA polymerase lambda diverged in evolution from DNA polymerasebeta toward specific Mn(++) dependence: A kinetic and thermodynamic study. Biochemistry 2003, 42, 7467–7476. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.J.; Garcia-Ortiz, M.V.; Esteban, V.; Blanco, L. Ribonucleotides and manganese ions improve non-homologous end joining by human Pol µ. Nucleic Acids Res. 2013, 41, 2428–2436. [Google Scholar] [CrossRef] [PubMed]
- Mourón, S.; Rodriguez-Acebes, S.; Martínez-Jiménez, M.I.; García-Gómez, S.; Chocrón, S.; Blanco, L.; Méndez, J. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Struct. Mol. Biol. 2013, 20, 1383–1389. [Google Scholar] [CrossRef] [PubMed]
- Stojkovič, G.; Makarova, A.V.; Wanrooij, P.H.; Forslund, J.; Burgers, P.M.; Wanrooij, S. Oxidative DNA damage stalls the human mitochondrial replisome. Sci. Rep. 2016, 6, a28942. [Google Scholar] [CrossRef] [PubMed]
- Badaracco, G.; Valsasnini, P.; Foiani, M.; Benfante, R.; Lucchini, G.; Plevani, P. Mechanism of initiation of in vitro DNA synthesis by the immunopurified complex between yeast DNA polymerase I and DNA primase. Eur. J. Biochem. 1986, 161, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Gronostajski, R.M.; Field, J.; Hurwitz, J. Purification of a primase activity associated with DNA polymerase alpha from HeLa cells. J. Biol. Chem. 1984, 259, 9479–9486. [Google Scholar] [PubMed]
- Tseng, B.Y.; Ahlem, C.N. DNA primase activity from human lymphocytes. Synthesis of oligoribonucleotides that prime DNA synthesis. J. Biol. Chem. 1982, 257, 7280–7283. [Google Scholar] [PubMed]
- Schiavone, D.; Jozwiakowski, S.K.; Romanello, M.; Guilbaud, G.; Guilliam, T.A.; Bailey, L.J.; Sale, J.E.; Doherty, A.J. PrimPol is Required for Replicative Tolerance of G Quadruplexes in Vertebrate Cells. Mol. Cell 2016, 61, 16–169. [Google Scholar] [CrossRef] [PubMed]
- Yang, W. An overview of Y-Family DNA polymerases and a case study of human DNA polymerase η. Biochemistry 2014, 53, 2793–2803. [Google Scholar] [CrossRef] [PubMed]
- Guilliam, T.A.; Brissett, N.C.; Ehlinger, A.; Keen, B.A.; Kolesar, P.; Taylor, E.M.; Bailey, L.J.; Lindsay, H.D.; Chazin, W.J.; Doherty, A.J. Molecular basis for PrimPol recruitment to replication forks by RPA. Nat. Commun. 2017, 8, 15222. [Google Scholar] [CrossRef] [PubMed]
- Bailey, L.J.; Bianchi, J.; Hégarat, N.; Hochegger, H.; Doherty, A.J. PrimPol-deficient cells exhibit a pronounced G2 checkpoint response following UV damage. Cell Cycle 2016, 15, 908–918. [Google Scholar] [CrossRef] [PubMed]
- Guilliam, T.A.; Bailey, L.J.; Brissett, N.C.; Doherty, A.J. PolDIP2 interacts with human PrimPol and enhances its DNA polymerase activities. Nucleic Acids Res. 2016, 44, 3317–3329. [Google Scholar] [CrossRef] [PubMed]
- Pilzecker, B.; Buoninfante, O.A.; Pritchard, C.; Blomberg, O.S.; Huijbers, I.J.; van den Berk, P.C.; Jacobs, H. PrimPol prevents APOBEC/AID family mediated DNA mutagenesis. Nucleic Acids Res. 2016, 44, 4734–4744. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Guilliam, T.A.; Tsuda, M.; Yamamoto, J.; Bailey, L.J.; Iwai, S.; Takeda, S.; Doherty, A.J.; Hirota, K. Repriming by PrimPol is critical for DNA replication restart downstream of lesions and chain-terminating nucleosides. Cell Cycle 2016, 15, 1997–2008. [Google Scholar] [CrossRef] [PubMed]
- Vallerga, M.B.; Mansilla, S.F.; Federico, M.B.; Bertolin, A.P.; Gottifredi, V. Rad51 recombinase prevents Mre11 nuclease-dependent degradation and excessive PrimPol-mediated elongation of nascent DNA after UV irradiation. Proc. Natl. Acad. Sci. USA 2015, 112, E6624–E6633. [Google Scholar] [CrossRef] [PubMed]
- Diamant, N.; Hendel, A.; Vered, I.; Carell, T.; Reissner, T.; de Wind, N.; Geacinov, N.; Livneh, Z. DNA damage bypass operates in the S and G2 phases of the cell cycle and exhibits differential mutagenicity. Nucleic Acids Res. 2012, 40, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Elvers, I.; Johansson, F.; Groth, P.; Erixon, K.; Helleday, T. UV stalled replication forks restart by re-priming in human fibroblasts. Nucleic Acids Res. 2011, 39, 7049–7057. [Google Scholar] [CrossRef] [PubMed]
- Guilliam, T.A.; Doherty, A.J. PrimPol-Prime Time to Reprime. Genes 2017, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Rodriguez-Belmonte, E.M.; Mazloum, N.; Xie, B.; Lee, M.Y. Identification of a novel protein, PDIP38, that interacts with the p50 subunit of DNA polymerase delta and proliferating cell nuclear antigen. J. Biol. Chem. 2003, 278, 10041–10047. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Kanki, T.; Fukuoh, A.; Ohgaki, K.; Takeya, R.; Aoki, Y.; Hamasaki, N.; Kang, D. PDIP38 associates with proteins constituting the mitochondrial DNA nucleoid. J. Biochem. 2005, 138, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Torregrosa-Muñumer, R.; Forslund, J.M.E.; Goffart, S.; Stojkovic, G.; Pfeiffer, A.; Carvalho, G.; Blanco, L.; Wanrooij, S.; Pohjoismäki, J.L.O.; et al. PrimPol is required for replication re-initiation after mitochondrial DNA damage. Proc. Natl. Acad. Sci. USA 2017. under review. [Google Scholar]
- Bharti, S.K.; Sommers, J.A.; Zhou, J.; Kaplan, D.L.; Spelbrink, J.N.; Mergny, J.L.; Brosh, R.M., Jr. DNA sequences proximal to human mitochondrial DNA deletion breakpoints prevalent in human disease form G-quadruplexes, a class of DNA structures inefficiently unwound by the mitochondrial replicative Twinkle helicase. J. Biol. Chem. 2014, 289, 29975–29993. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.W.; Pereira, F.; Barrett, S.P.; Kolesar, J.E.; Cao, K.; Damas, J.; Yatsunyk, L.A.; Johnson, F.B.; Kaufman, B.A. Association of G-quadruplex forming sequences with human mtDNA deletion breakpoints. BMC Genom. 2014, 15, a677. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.C.; Tseng, T.Y.; Chen, Y.T.; Chang, C.C.; Wang, Z.F.; Wang, C.L.; Hsu, T.N.; Li, P.T.; Chen, C.T.; Lin, J.J.; et al. Direct evidence of mitochondrial G-quadruplex DNA by using fluorescent anti-cancer agents. Nucleic Acids Res. 2015, 43, 10102–10113. [Google Scholar] [CrossRef] [PubMed]
- Wanrooij, P.H.; Uhler, J.P.; Shi, Y.; Westerlund, F.; Falkenberg, M.; Gustafsson, C.M. A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop. Nucleic Acids Res. 2012, 40, 10334–10344. [Google Scholar] [CrossRef] [PubMed]
- Wanrooij, P.H.; Uhler, J.P.; Simonsson, T.; Falkenberg, M.; Gustafsson, C.M. G-quadruplex structures in RNA stimulate mitochondrial transcription termination and primer formation. Proc. Natl. Acad. Sci. USA 2010, 107, 16072–16077. [Google Scholar] [CrossRef] [PubMed]
- Kelman, Z. PCNA: Structure, functions and interactions. Oncogene 1997, 14, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Makarova, A.V.; Stodola, J.L.; Burgers, P.M. A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012, 40, 11618–11626. [Google Scholar] [CrossRef] [PubMed]
- Masuda, Y.; Kanao, R.; Kaji, K.; Ohmori, H.; Hanaoka, F.; Masutani, C. Different types of interaction between PCNA and PIP boxes contribute to distinct cellular functions of Y-family DNA polymerases. Nucleic Acids Res. 2015, 43, 7898–7910. [Google Scholar] [CrossRef] [PubMed]
- Vidal, A.E.; Kannouche, P.; Podust, V.N.; Yang, W.; Lehmann, A.R.; Woodgate, R. Proliferating cell nuclear antigen-dependent coordination of the biological functions of human DNA polymerase iota. J. Biol. Chem. 2004, 279, 48360–48368. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Jiménez, M.I.; Lahera, A.; Blanco, L. Human PrimPol activity is enhanced by RPA. Sci. Rep. 2017, 7, a783. [Google Scholar] [CrossRef] [PubMed]
- Maga, G.; Crespan, E.; Markkanen, E.; Imhof, R.; Furrer, A.; Villani, G.; Hübscher, U.; van Loon, B. DNA polymerase δ-interacting protein 2 is a processivity factor for DNA polymerase λ during 8-oxo-7,8-dihydroguanine bypass. Proc. Natl. Acad. Sci. USA 2013, 110, 18850–18855. [Google Scholar] [CrossRef] [PubMed]
- Tissier, A.; Janel-Bintz, R.; Coulon, S.; Klaile, E.; Kannouche, P.; Fuchs, R.P.; Cordonnier, A.M. Crosstalk between replicative and translesional DNA polymerases: PDIP38 interacts directly with Pol eta. DNA Repair 2010, 9, 922–928. [Google Scholar] [CrossRef] [PubMed]
- Klaile, E.; Müller, M.M.; Kannicht, C.; Otto, W.; Singer, B.B.; Reutter, W.; Obrink, B.; Lucka, L. The cell adhesion receptor carcinoembryonic antigen-related cell adhesion molecule 1 regulates nucleocytoplasmic trafficking of DNA polymerase δ-interacting protein 38. J. Biol. Chem. 2007, 282, 26629–26640. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Li, H.; Wang, Q.; Xie, S.; Rahmeh, A.; Dai, W.; Lee, M.Y. Further characterization of human DNA polymerase δ interacting protein 38. J. Biol. Chem. 2005, 280, 22375–22384. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Wu, J.; Xue, A.; Su, Y.; Wang, X.; Lu, X.; Zhou, Z.; Qu, J.; Zhou, X. Exome sequencing reveals CCDC111 mutation associated with high myopia. Hum. Genet. 2013, 132, 913–921. [Google Scholar] [CrossRef] [PubMed]
- Keen, B.A.; Bailey, L.J.; Jozwiakowski, S.K.; Doherty, A.J. Human PrimPol mutation associated with high myopia has a DNA replication defect. Nucleic Acids Res. 2014, 42, 12102–12111. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, Q. PRIMPOL mutation: Functional study does not always reveal the truth. Investig. Ophthalmol. Vis. Sci. 2015, 56, 1181–1182. [Google Scholar] [CrossRef] [PubMed]
- Roos, W.P.; Tsaalbi-Shtylik, A.; Tsaryk, R.; Güvercin, F.; de Wind, N.; Kaina, B. The translesion polymerase Rev3L in the tolerance of alkylating anticancer drugs. Mol. Pharmacol. 2009, 76, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Lin, X.; Okuda, T.; Howell, S.B. DNA polymerase zeta regulates cisplatin cytotoxicity, mutagenicity, and the rate of development of cisplatin resistance. Cancer Res. 2004, 64, 8029–8035. [Google Scholar] [CrossRef] [PubMed]

| DNA Damage | PrimPol TLS In Vitro | |
|---|---|---|
| oxidative lesions | 8-oxo-G | - bypasses 8-oxo-G incorporating dATP and dCTP with equal efficiency [26,27]; - bypasses 8-oxo-G and preferentially incorporates dC [33,42] |
| TG (thymidine glycol) | - does not bypass TG [26,35]; - PrimPol 1–354 incorporates a nucleotide opposite TG but cannot extend from the lesion [35] | |
| photo-products | cis-syn T–T dimers | - bypasses CPD cis-syn T–T dimers [41]; - does not bypass cis-syn T–T dimers but extends a primer terminus with two dA residues annealed opposite the T–T CPD [26]; - PrimPol1-354 bypasses cis-syn T–T dimers with high efficiency and fidelity [35] |
| T–T (6–4) photoproducts | - bypasses T–T (6–4) photoproducts in error-prone manner incorporating dTTP opposite 3′Т and dGTP/dCTP opposite 5′Т [26] or by skipping mechanism [27,34,41] | |
| abasic sites | - does not bypass lesion [26,35,42]; - bypasses lesion with high efficiency using skipping mechanism [27,34]; - bypasses with very weak efficiency and shows nearly equal preference for either skipping the abasic site or inserting dAMP [33] | |
| deoxyuracil | - bypasses as T and incorporates dATP opposite the lesion [35] | |
| ▪ Protein-Partner | Localization of Protein in DNA Compartment | Effect on PrimPol Activity |
|---|---|---|
| ▪ RPA (replication protein A) | nuclear | inhibits primase and polymerase activities on short DNA templates [30,42,69] but stimulates primase and polymerase activities on long DNA templates when non-saturating in vitro [48,69], targets PrimPol to DNA damage sites in nuclei in vivo [28,30,48] |
| ▪ mtSSB (mitochondrial single-stranded DNA-binding protein) | mitochondrial | inhibits primase and polymerase activities on short DNA templates in vitro [42] |
| ▪ PolDIP2 (polymerase delta-interacting protein 2) | mitochondrial and possibly nuclear | stimulates DNA polymerase activity in vitro [50] |
| ▪ Twinkle | mitochondrial | stimulates DNA polymerase activity in vitro [42] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boldinova, E.O.; Wanrooij, P.H.; Shilkin, E.S.; Wanrooij, S.; Makarova, A.V. DNA Damage Tolerance by Eukaryotic DNA Polymerase and Primase PrimPol. Int. J. Mol. Sci. 2017, 18, 1584. https://doi.org/10.3390/ijms18071584
Boldinova EO, Wanrooij PH, Shilkin ES, Wanrooij S, Makarova AV. DNA Damage Tolerance by Eukaryotic DNA Polymerase and Primase PrimPol. International Journal of Molecular Sciences. 2017; 18(7):1584. https://doi.org/10.3390/ijms18071584
Chicago/Turabian StyleBoldinova, Elizaveta O., Paulina H. Wanrooij, Evgeniy S. Shilkin, Sjoerd Wanrooij, and Alena V. Makarova. 2017. "DNA Damage Tolerance by Eukaryotic DNA Polymerase and Primase PrimPol" International Journal of Molecular Sciences 18, no. 7: 1584. https://doi.org/10.3390/ijms18071584
APA StyleBoldinova, E. O., Wanrooij, P. H., Shilkin, E. S., Wanrooij, S., & Makarova, A. V. (2017). DNA Damage Tolerance by Eukaryotic DNA Polymerase and Primase PrimPol. International Journal of Molecular Sciences, 18(7), 1584. https://doi.org/10.3390/ijms18071584