Deep Circular RNA Sequencing Provides Insights into the Mechanism Underlying Grass Carp Reovirus Infection
Abstract
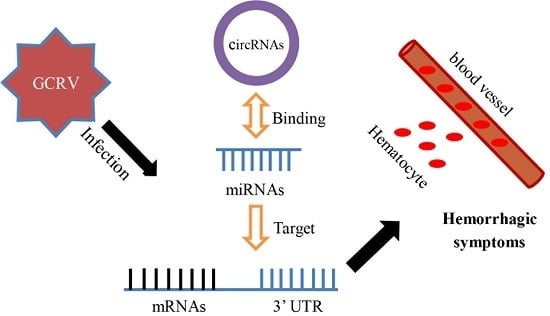
:1. Introduction
2. Results
2.1. Preliminary Analysis of circRNA Sequencing
2.2. Identification of circRNAs before and after GCRV Infection
2.3. Differentially Expressed circRNAs Following GCRV Infection
2.4. Characterisation of Parental Genes of Differentially Expressed circRNAs
2.5. Prediction of Binding miRNAs of Differentially Expressed circRNAs
2.6. Confirmation of circRNAs by PCR and RT-qPCR
3. Discussion
4. Materials and Methods
4.1. Ethics Approval and Consent to Participate
4.2. Experimental Fish
4.3. Virus Challenge and Sample Collection
4.4. RNA Isolation, Library Construction, and Sequencing
4.5. Data Analysis
4.6. Differential Expression Analysis and Binding miRNA Prediction
4.7. PCR Amplification and Sanger Sequencing
4.8. Validation of circRNA and Parental Gene Expression Level by RT-qPCR
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| APOA4 | Apolipo A-IV-like |
| BCV | Biological coefficient of variation |
| C3 | Complement component 3 |
| CD84 | SLAM family member 5 |
| circRNAs | Circular RNAs |
| ciRNAs | Circular intronic RNAs |
| CXCR-4 | C-X-C chemokine receptor type 4 |
| dpi | Days post-infection |
| ecircRNAs | Circular exonic RNAs |
| eiciRNAs | Exon-intron circRNAs |
| F13A | Coagulation factor XIII A chain |
| GCRV | Grass carp reovirus |
| HBAC | Hemoglobin cathodic subunit α |
| HERC1 | Probable E3 ubiquitin-protein ligase HERC1 |
| HERC4 | Probable E3 ubiquitin-protein ligase HERC4 |
| ITGA8 | Integrin α-8 |
| ITIH6 | Inter-α-trypsin inhibitor heavy chain H6-like |
| KAT6B | Histone acetyltransferase KAT6B |
| KCNC1 | Potassium voltage-gated channel subfamily C member 1 |
| MIB1 | E3 ubiquitin-protein ligase mib1 |
| PBS | Phosphate buffered solution |
| Pre-mRNA | mRNA precursors |
| SLAM | Signaling lymphocytic activation molecule |
| TPM | Transcripts per million |
References
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Starke, S.; Jost, I.; Rossbach, O.; Schneider, T.; Schreiner, S.; Hung, L.H.; Bindereif, A. Exon circularization requires canonical splice signals. Cell Rep. 2015, 10, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Nigro, J.M.; Cho, K.R.; Fearon, E.R.; Kern, S.E.; Ruppert, J.M.; Oliner, J.D.; Kinzler, K.W.; Vogelstein, B. Scrambled exons. Cell 1991, 64, 607–613. [Google Scholar] [CrossRef]
- Capel, B.; Swain, A.; Nicolis, S.; Hacker, A.; Walter, M.; Koopman, P.; Goodfellow, P.; Lovell-Badge, R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993, 73, 1019–1030. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed]
- Rybak-Wolf, A.; Stottmeister, C.; Glazar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Guo, X.; Wang, W. Identification and characterization of circular RNAs in zebrafish. FEBS Lett. 2017, 591, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Petkovic, S.; Müller, S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015, 43, 2454–2465. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Guo, J.; Chen, Y.; Chang, C.; Xu, C. Comprehensive CircRNA expression profile and selection of key CircRNAs during priming phase of rat liver regeneration. BMC Genom. 2017, 18, 80. [Google Scholar] [CrossRef] [PubMed]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef] [PubMed]
- Kulcheski, F.R.; Christoff, A.P.; Margis, R. Circular RNAs are miRNA sponges and can be used as a new class ofbiomarker. J. Biotechnol. 2016, 238, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Cortés-López, M.; Miura, P. Emerging functions of circular RNAs. Yale J. Biol. Med. 2016, 89, 527–537. [Google Scholar] [PubMed]
- Tang, C.M.; Zhang, M.; Huang, L.; Hu, Z.Q.; Zhu, J.N.; Xiao, Z.; Zhang, Z.; Lin, Q.X.; Zheng, X.L.; Yang, M.; et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci. Rep. 2017, 7, 40342. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.N.; Li, J.; Zhu, C.L.; Feng, W.T.; Shao, J.X.; Wan, L.; Huang, M.D.; He, J.D. Comprehensive profile of differentially expressed circular RNAs reveals thathsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. OncoTargets Ther. 2016, 9, 7451–7458. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, Y.; Zheng, Q.; Bao, C.; He, J.; Chen, B.; Lyu, D.; Zheng, B.; Xu, Y.; Long, Z.; et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017, 388, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Feng, C.Y.; Xiang, Z.; Chen, Y.P.; Li, Y.M. CircRNA expression pattern and circRNA-miRNA -mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget 2016, 7, 66455–66467. [Google Scholar] [CrossRef] [PubMed]
- Nan, A.; Chen, L.; Zhang, N.; Liu, Z.; Yang, T.; Wang, Z.; Yang, C.; Jiang, Y. A novel regulatory network among LncRpa, CircRar1, MiR-671 and apoptotic genes promotes lead-induced neuronal cell apoptosis. Arch. Toxicol. 2017, 91, 1671–1684. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization (FAO). Fishery and Aquaculture Statistics Yearbook; Food and Agriculture Organization of the United Nations: Rome, Italy, 2016. [Google Scholar]
- Rao, Y.; Su, J. Insights into the antiviral immunity against grass carp (Ctenopharyngodon idella) reovirus (GCRV) in grass carp. J. Immunol. Res. 2015, 2015, 670437. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zeng, W.; Liu, C.; Zhang, C.; Wang, Y.; Shi, C.; Wu, S. Complete genome sequence of a reovirus isolated from grass carp, indicating different genotypes of GCRV in China. J. Virol. 2012, 86, 12466. [Google Scholar] [CrossRef] [PubMed]
- Jian, J.C.; Wang, Y.; Yan, X.Y.; Ding, Y.; Wu, Z.H.; Lu, Y.S. Molecular cloning and prokaryotic expression of vp5 gene of grass carp reovirus strain GCRV096. Virus Genes 2013, 47, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Fan, Y.D.; Zeng, L.B.; Ma, J. Prokaryotic expression and immunoassay of grass carp reovirus capsid VP6 protein. Acta Virol. 2013, 57, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.L.; Zhang, L.F.; Fang, Z.Z.; Xu, L.P.; Zhang, M.; Wang, N.; Jiang, Y.L.; Lin, X.M. Detection of grass carp reovirus (GCRV) with monoclonal antibodies. Arch Virol. 2014, 159, 649–655. [Google Scholar]
- Zeng, W.; Wang, Y.; Liang, H.; Liu, C.; Song, X.; Shi, C.; Wu, S.; Wang, Q. A one-step duplex rRT-PCR assay for the simultaneous detection of grass carp reovirus genotypes I and II. J. Virol. Methods 2014, 210, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Liu, G.L.; Gong, Y.X.; Ling, F.; Wang, G.X. Protective immunity of grass carp immunized with DNA vaccine encoding the vp7 gene of grass carp reovirus using carbon nanotubes as a carrier molecule. Fish Shellfish Immunol. 2015, 42, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Song, L.; Wang, H.; Xu, X.; Wang, T.; Lu, L. Proteomic analysis of cellular protein expression profiles in response to grass carp reovirus infection. Fish Shellfish Immunol. 2015, 44, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhang, A.; Chu, P.; Li, Y.; Huang, R.; Liao, L.; Zhu, Z.; Wang, Y. Deep Illumina sequencing reveals conserved and novel microRNAs in grass carpin response to grass carp reovirus infection. BMC Genom. 2017, 18, 195. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Xiao, S.; Qiu, C.; Wang, Z. Transcriptome-wide identification and functional investigation of circular RNA in the teleost large yellow croaker (Larimichthys crocea). Mar. Genom. 2017, 32, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Cui, L.; Zhou, Y.; Zhu, C.; Fan, D.; Gong, H.; Zhao, Q.; Zhou, C.; Zhao, Y.; Lu, D.; et al. Transcriptome-wide investigation of circular RNAs in rice. RNA 2015, 21, 2076–2087. [Google Scholar] [CrossRef] [PubMed]
- Danan, M.; Schwartz, S.; Edelheit, S.; Sorek, R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012, 40, 3131–3142. [Google Scholar] [CrossRef] [PubMed]
- Subramanian Vignesh, K.; Deepe, G.S., Jr. Immunological orchestration of zinc homeostasis: The battle between host mechanisms and pathogen defenses. Arch. Biochem. Biophys. 2016, 611, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Jesse, H.E.; Roberts, I.S.; Cavet, J.S. Metal ion homeostasis in Listeria monocytogenes and importance in host-pathogen interactions. Adv. Microb. Physiol. 2014, 65, 83–123. [Google Scholar] [PubMed]
- Hochrainer, K.; Mayer, H.; Baranyi, U.; Binder, B.; Lipp, J.; Kroismayr, R. The human HERC family of ubiquitin ligases: Novel members, genomic organization, expression profiling, and evolutionary aspects. Genomics 2005, 85, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007, 23, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhang, A.; Pei, Y.; Chu, P.; Li, Y.; Huang, R.; Liao, L.; Zhu, Z.; Wang, Y. Differences in responses of grass carp to different types of grass carp reovirus(GCRV) and the mechanism of hemorrhage revealed by transcriptome sequencing. BMC Genom. 2017, 18, 452. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions andgene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Lu, Y.; Zhang, Y.; Ning, Z.; Li, Y.; Zhao, Q.; Lu, H.; Huang, R.; Xia, X.; Feng, Q.; et al. The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat. Genet. 2015, 47, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Chen, J.; Li, Z.; Li, X.; Hu, X.; Huang, Y.; Zhao, X.; Liang, C.; Wang, Y.; Sun, L.; et al. Integrated profiling of microRNAs and mRNAs: MicroRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS ONE 2010, 5, e15224. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]








| Sample | Duplicates | Raw Reads | Clean Reads | Clean Bases (Gb) | Error Rate | Q20 | Q30 | GC Content |
|---|---|---|---|---|---|---|---|---|
| C | C-a | 70,897,548 | 61,922,082 | 9.28 | 0.03 | 97.00 | 92.97 | 67.28 |
| C-b | 84,758,120 | 75,871,992 | 11.38 | 0.03 | 97.34 | 93.48 | 65.24 | |
| C-c | 71,105,834 | 67,836,022 | 10.18 | 0.05 | 94.06 | 86.95 | 65.86 | |
| T1 | T1-a | 70,155,734 | 65,647,992 | 9.84 | 0.04 | 95.15 | 89.03 | 69.19 |
| T1-b | 66,689,052 | 62,852,844 | 9.42 | 0.04 | 95.16 | 88.92 | 67.40 | |
| T1-c | 72,041,160 | 68,785,204 | 10.32 | 0.04 | 95.08 | 88.81 | 65.47 | |
| T3 | T3-a | 72,188,758 | 67,758,712 | 10.16 | 0.04 | 95.17 | 89.05 | 67.40 |
| T3-b | 73,758,060 | 68,452,540 | 10.26 | 0.03 | 96.00 | 90.70 | 67.08 | |
| T3-c | 70,612,416 | 66,350,154 | 9.96 | 0.03 | 96.27 | 91.04 | 64.72 | |
| T5 | T5-a | 89,647,100 | 83,873,758 | 12.58 | 0.03 | 96.30 | 91.03 | 64.46 |
| T5-b | 79,912,480 | 74,507,152 | 11.18 | 0.03 | 96.46 | 91.38 | 64.51 | |
| T5-c | 87,519,928 | 82,623,624 | 12.40 | 0.03 | 96.49 | 91.56 | 64.62 | |
| T7 | T7-a | 67,055,966 | 62,921,544 | 9.44 | 0.03 | 96.27 | 91.28 | 65.49 |
| T7-b | 74,675,616 | 69,963,696 | 10.50 | 0.03 | 96.55 | 91.70 | 64.21 | |
| T7-c | 67,217,152 | 62,582,180 | 9.38 | 0.03 | 96.35 | 91.35 | 65.14 |
| Comparison | Upregulated | Downregulated | Total |
|---|---|---|---|
| T1/C | 4 | 7 | 11 |
| T3/C | 8 | 12 | 20 |
| T5/C | 7 | 11 | 18 |
| T7/C | 9 | 8 | 17 |
| circRNAs | Parental Genes | Gene Description | Possible Function |
|---|---|---|---|
| cid_circ_0236 | MIB1 | E3 ubiquitin-protein ligase mib1 | metal ion binding |
| cid_circ_0735 | KAT6B | Histone acetyltransferase KAT6B | metal ion binding |
| cid_circ_1680 | ITGA8 | Integrin α-8 | metal ion binding |
| cid_circ_1941 | EZH1 | Histone-lysine N-methyltransferase EZH1 | metal ion binding |
| cid_circ_2374 | SUCLG2 | Succinyl-CoA ligasesubunit β, mitochondrial (Fragment) | metal ion binding |
| cid_circ_3014 | NLK | Serine/threonine-protein kinase NLK | metal ion binding |
| cid_circ_3204 | HBAC | Hemoglobin cathodic subunit α | metal ion binding |
| cid_circ_4439 | KCNC1 | Potassium voltage-gated channel subfamily C member 1 | metal ion binding |
| cid_circ_4698 | F13A | Coagulation factor XIII A chain | metal ion binding |
| cid_circ_1307 | HERC1 | Probable E3 ubiquitin-protein ligase HERC1 | protein ubiquitination |
| cid_circ_3884 | HERC4 | Probable E3 ubiquitin-protein ligase HERC4 | protein ubiquitination |
| cid_circ_1448 | UBP45 | Ubiquitin carboxyl-terminal hydrolase 45 | protein ubiquitination |
| cid_circ_4756 | ABTB2 | Ankyrin repeat and BTB/POZ domain-containing protein 2 | protein ubiquitination |
| cid_circ_0377 | AGPAT5 | 1-Acyl-sn-glycerol-3-phosphate acyltransferase | enzyme activity |
| cid_circ_1991 | SPAG9 | C-jun-amino-terminal kinase-interacting protein 4 | enzyme activity |
| cid_circ_3254 | PSTA | Phosphate transport system permease protein PstA | enzyme activity |
| cid_circ_3323 | GIMAP2 | GTPase IMAP family member 2 | enzyme activity |
| cid_circ_4000 | SLC12A2 | Solute carrier family 12 member 2 | enzyme activity |
| cid_circ_4579 | SRGAP3 | SLIT-ROBO Rho GTPase-activating protein 3 | enzyme activity |
| cid_circ_1226 | CELF1 | CUGBP Elav-like family member 1 | nucleotide binding |
| cid_circ_3806 | MRPL39 | 39S ribosomal protein L39, mitochondrial | nucleotide binding |
| cid_circ_3837 | ELF2 | ETS-related transcription factor Elf-2 | nucleotide binding |
| cid_circ_0698 | ABCD1 | ATP-binding cassette sub-family D member 1 | nucleotide-binding |
| cid_circ_1133 | LAMA5 | Laminin subunit α-5 | other |
| cid_circ_1967 | TANC2 | Protein TANC2 | other |
| cid_circ_2040 | TMEM187 | Transmembrane protein 187 | other |
| cid_circ_2063 | PHYIP | Phytanoyl-CoA hydroxylase-interacting protein | other |
| cid_circ_2263 | GRN | Granulin-1 | other |
| cid_circ_3408 | DMTN | Dematin | other |
| cid_circ_4314 | ASPP1 | Apoptosis-stimulating of p53 protein 1 | other |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, L.; Zhang, A.; Xiong, L.; Li, Y.; Huang, R.; Liao, L.; Zhu, Z.; Wang, A.Y. Deep Circular RNA Sequencing Provides Insights into the Mechanism Underlying Grass Carp Reovirus Infection. Int. J. Mol. Sci. 2017, 18, 1977. https://doi.org/10.3390/ijms18091977
He L, Zhang A, Xiong L, Li Y, Huang R, Liao L, Zhu Z, Wang AY. Deep Circular RNA Sequencing Provides Insights into the Mechanism Underlying Grass Carp Reovirus Infection. International Journal of Molecular Sciences. 2017; 18(9):1977. https://doi.org/10.3390/ijms18091977
Chicago/Turabian StyleHe, Libo, Aidi Zhang, Lv Xiong, Yongming Li, Rong Huang, Lanjie Liao, Zuoyan Zhu, and And Yaping Wang. 2017. "Deep Circular RNA Sequencing Provides Insights into the Mechanism Underlying Grass Carp Reovirus Infection" International Journal of Molecular Sciences 18, no. 9: 1977. https://doi.org/10.3390/ijms18091977
APA StyleHe, L., Zhang, A., Xiong, L., Li, Y., Huang, R., Liao, L., Zhu, Z., & Wang, A. Y. (2017). Deep Circular RNA Sequencing Provides Insights into the Mechanism Underlying Grass Carp Reovirus Infection. International Journal of Molecular Sciences, 18(9), 1977. https://doi.org/10.3390/ijms18091977