Genome-Wide Identification of the PHD-Finger Family Genes and Their Responses to Environmental Stresses in Oryza sativa L.
Abstract
:1. Introduction
2. Results
2.1. Identification and Chromosomal Location of the Rice PHD Family Genes
2.2. Phylogenetic Analyses and Gene Architecture of the Rice PHD Family
2.3. Investigation of the Conserved Functional Domains in OsPHD Proteins
2.4. Expression Profiles of OsPHD Genes in Rice Tissues
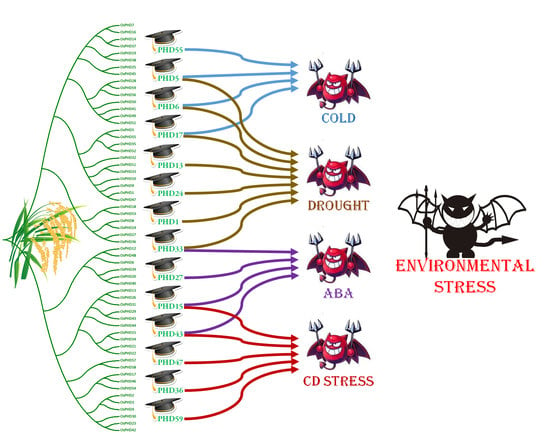
2.5. Expression Responses of OsPHD Genes to Environmental Stresses
2.6. Quantitative Real-Time PCR Analyses of OsPHDs Expression in Response to Environmental Stresses
3. Discussion
4. Materials and Methods
4.1. Identification of the PHD-Finger Family Genes in Rice
4.2. Multiple Alignment, Phylogenetic Analysis and Chromosomal Location of the Rice PHD Family Genes
4.3. Analyses of Gene Structure, Functional Domains and Conserved Motifs of the Rice PHD Family
4.4. Expression Analysis of the Rice PHD Family Genes in Diverse Tissues and Under Different Stress Treatments
4.5. Quantitative Real-Time PCR Analyses of the Rice PHD Family Genes in Response to Environmental Stresses
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Almeida, D.M.; Almadanim, M.C.; Lourenco, T.; Abreu, I.A.; Saibo, N.J.; Oliveira, M.M. Screening for abiotic stress tolerance in rice: Salt, cold, and drought. Methods Mol. Biol. 2016, 1398, 155–182. [Google Scholar] [PubMed]
- Zhang, Q.; Chen, Q.; Wang, S.; Hong, Y.; Wang, Z. Rice and cold stress: Methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice 2014, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Hadiarto, T.; Tran, L.S.P. Progress studies of drought-responsive genes in rice. Plant Cell Rep. 2011, 30, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.L.; Sun, C.D.; Shen, C.J.; Wang, S.K.; Liu, F.; Liu, Y.; Chen, Y.L.; Li, C.Y.; Qian, Q.; Aryal, B.; et al. The auxin transporter, OsAUX1, is involved in primary root and root hair elongation and in Cd stress responses in rice (Oryza sativa L.). Plant J. 2015, 83, 818–830. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Huangfu, J.; Li, J.; Li, R.; Ye, M.; Kuai, P.; Zhang, T.; Lou, Y. The transcription factor OsWRKY45 negatively modulates the resistance of rice to the brown planthopper Nilaparvata lugens. Int. J. Mol. Sci. 2016, 17, 697. [Google Scholar] [CrossRef] [PubMed]
- Jaffar, M.A.; Song, A.; Faheem, M.; Chen, S.; Jiang, J.; Liu, C.; Fan, Q.; Chen, F. Involvement of CmWRKY10 in drought tolerance of chrysanthemum through the ABA-signaling pathway. Int. J. Mol. Sci. 2016, 17, 693. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Jones, D.C.; Li, W.; Xie, F.; Ma, J.; Sun, R.; Wang, Q.; Zhu, S.; Zhang, B. Genome-wide identification of R2R3-MYB genes and expression analyses During Abiotic Stress in Gossypium raimondii. Sci. Rep. 2016, 6, 22980. [Google Scholar] [CrossRef] [PubMed]
- Thao, N.P.; Thu, N.B.A.; Hoang, X.L.T.; Ha, C.V.; Tran, L.P. Differential expression analysis of a subset of drought-responsive GmNAC genes in two soybean cultivars differing in drought tolerance. Int. J. Mol. Sci. 2013, 14, 23828–23841. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Lv, B.; Luo, L.; He, J.; Mao, C.; Xi, D.; Ming, F. The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci. Rep. 2017, 7, 40641. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Roychoudhury, A. Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 2017, 254, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Arora, R.; Ray, S.; Singh, A.K.; Singh, V.P.; Takatsuji, H.; Kapoor, S.; Tyagi, A.K. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 2007, 65, 467–485. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, D.; Dejardin, A.; Leple, J.C.; Lesage-Descauses, M.C.; Pilate, G. Genome-wide analysis of LIM gene family in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa. DNA Res. 2007, 14, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Takatsuji, H. Zinc-finger transcription factors in plants. Cell Mol. Life Sci. 1998, 54, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Aasland, R.; Gibson, T.J.; Stewart, A.F. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 1995, 20, 56–59. [Google Scholar] [CrossRef]
- Borden, K.L.; Freemont, P.S. The RING finger domain: A recent example of a sequence-structure family. Curr. Opin. Struct. Biol. 1996, 6, 395–401. [Google Scholar] [CrossRef]
- Schindler, U.; Beckmann, H.; Cashmore, A.R. HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 1993, 4, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Liu, Q.P.; Xue, Q.Z. Comparative phylogenetic analysis of the rice and Arabidopsis PHD-finger proteins. Acta Genet. Sin. 2004, 31, 1284–1293. [Google Scholar] [PubMed]
- Wu, S.; Wu, M.; Dong, Q.; Jiang, H.; Cai, R.; Xiang, Y. Genome-wide identification, classification and expression analysis of the PHD-finger protein family in Populus trichocarpa. Gene 2015, 575, 75. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, J.; Wang, Y.; Zhao, Y.; Jiang, H.; Cheng, B. Systematic analysis of the maize PHD-finger gene family reveals a subfamily involved in abiotic stress response. Int. J. Mol. Sci. 2015, 16, 23517–23544. [Google Scholar] [CrossRef] [PubMed]
- Molitor, A.M.; Bu, Z.; Yu, Y.; Shen, W.H. Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PLoS Genet. 2014, 10, e1004091. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Gonzalez, L.; Mouriz, A.; Narro-Diego, L.; Bustos, R.; Martinez-Zapater, J.M.; Jarillo, J.A.; Pineiro, M. Chromatin-dependent repression of the Arabidopsis floral integrator genes involves plant specific PHD-containing proteins. Plant Cell 2014, 26, 3922–3938. [Google Scholar] [CrossRef] [PubMed]
- Fernandez Gomez, J.; Wilson, Z.A. A barley PHD finger transcription factor that confers male sterility by affecting tapetal development. Plant Biotechnol. J. 2014, 12, 765–777. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Huang, J.; Hao, Y.J.; Zou, H.F.; Wang, H.W.; Zhao, J.Y.; Liu, X.Y.; Zhang, W.K.; Ma, B.; Zhang, J.S.; et al. Soybean GmPHD-type transcription regulators improve stress tolerance in transgenic Arabidopsis plants. PLoS ONE 2009, 4, e7209. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Pi, E.X.; Tsai, S.N.; Lam, H.M.; Sun, S.M.; Kwan, Y.W.; Ngai, S.M. GmPHD5 acts as an important regulator for crosstalk between histone H3K4 di-methylation and H3K14 acetylation in response to salinity stress in soybean. BMC Plant Biol. 2011, 11, 178. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Gong, Z.; Lu, X.; Miao, D.; Shi, J.; Lu, J.; Zhao, Y. Germostatin resistance locus 1 encodes a PHD finger protein involved in auxin-mediated seed dormancy and germination. Plant J. 2016, 85, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Chujo, A.; Eiguchi, M.; Kurata, N. Radial axis differentiation in a globular embryo is marked by HAZ1, a PHD-finger homeobox gene of rice. Gene 2004, 331, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Yamanouchi, U.; Nonoue, Y.; Sugimoto, K.; Wang, Z.X.; Minobe, Y.; Yano, M. Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J. 2011, 66, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yuan, Z.; Vizcay-Barrena, G.; Yang, C.; Liang, W.; Zong, J.; Wilson, Z.A.; Zhang, D. PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 2011, 156, 615–630. [Google Scholar] [CrossRef] [PubMed]
- Winicov, I.I.; Bastola, D.R. Transgenic overexpression of the transcription factor alfin1 enhances expression of the endogenous MsPRP2 gene in alfalfa and improves salinity tolerance of the plants. Plant Physiol. 1999, 120, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Nagata, N.; Yoshiba, Y.; Ohme-Takagi, M.; Ma, H.; Shinozaki, K. Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 2007, 19, 3549–3562. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Shinozaki, K. The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant Cell Physiol. 2002, 43, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Wilson, Z.A.; Morroll, S.M.; Dawson, J.; Swarup, R.; Tighe, P.J. The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 2001, 28, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Reddy, T.V.; Kaur, J.; Agashe, B.; Sundaresan, V.; Siddiqi, I. The DUET gene is necessary for chromosome organization and progression during male meiosis in Arabidopsis and encodes a PHD finger protein. Development 2003, 130, 5975–5987. [Google Scholar] [CrossRef] [PubMed]
- Greb, T.; Mylne, J.S.; Crevillen, P.; Geraldo, N.; An, H.; Gendall, A.R.; Dean, C. The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr. Biol. 2007, 17, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Mussig, C.; Kauschmann, A.; Clouse, S.D.; Altmann, T. The Arabidopsis PHD-finger protein SHL is required for proper development and fertility. Mol. Gen. Genet. 2000, 264, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Winicov, I. Alfin1 transcription factor overexpression enhances plant root growth under normal and saline conditions and improves salt tolerance in alfalfa. Planta 2000, 210, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Callebaut, I.; Courvalin, J.C.; Mornon, J.P. The BAH (bromo-adjacent homology) domain: A link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 1999, 446, 189–193. [Google Scholar] [CrossRef]
- Oliver, A.W.; Jones, S.A.; Roe, S.M.; Matthews, S.; Goodwin, G.H.; Pearl, L.H. Crystal structure of the proximal BAH domain of the polybromo protein. Biochem. J. 2005, 389, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; De Vivo, I.; Slany, R.; Miyamoto, A.; Firestein, R.; Cleary, M.L. Association of SET domain and myotubularin-related proteins modulates growth control. Nat. Genet. 1998, 18, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Baumbusch, L.O.; Thorstensen, T.; Krauss, V.; Fischer, A.; Naumann, K.; Assalkhou, R.; Schulz, I.; Reuter, G.; Aalen, R.B. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001, 29, 4319–4333. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Min, J.R. Structure and function of the nucleosome-binding PWWP domain. Trends Biochem. Sci. 2014, 39, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Stec, I.; Nagl, S.B.; van Ommen, G.J.B.; den Dunnen, J.T. The PWWP domain: A potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 2000, 473, 1–5. [Google Scholar] [CrossRef]
- Chandrika, N.N.; Sundaravelpandian, K.; Schmidt, W. A PHD in histone language: On the role of histone methylation in plant responses to phosphate deficiency. Plant Sign. Behav. 2013, 8, e24381. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Wang, Z.; Liu, F.; Li, Z.; Peng, Q.; Guo, J.; Xu, L.; Que, Y. Isolation and characterization of ScGluD2, a newsugarcane β-1,3-Glucanase D Family gene induced by Sporisorium scitamineum, ABA, H2O2, NaCl, and CdCl2 stresses. Front. Plant Sci. 2016, 7, 1348. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Baek, W.; Jung, J.; Kim, J.; Lee, S.C. Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 2015, 16, 15251–15270. [Google Scholar] [CrossRef] [PubMed]
- Lindemose, S.; O’Shea, C.; Jensen, M.K.; Skriver, K. Structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 2013, 14, 5842–5878. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Hou, Y.; Wang, Y.; Li, Z.; Zhao, J.; Tong, X.; Lin, H.; Wei, X.; Ao, H.; Zhang, J. A comprehensive proteomic survey of ABA-induced protein phosphorylation in rice (Oryza sativa L.). Int. J. Mol. Sci. 2017, 18, 60. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, Y.; Yang, H.; Wang, W.; Wu, J.; Hu, X. Quantitative proteomic analyses identify ABA-related proteins and signal pathways in maize leaves under drought conditions. Front. Plant Sci. 2016, 7, 1827. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Tang, H.; Wang, X.; Paterson, A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2013, 41, D1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Takehisa, H.; Kamatsuki, K.; Minami, H.; Namiki, N.; Ikawa, H.; Ohyanagi, H.; Sugimoto, K.; Antonio, B.A.; Nagamura, Y. RiceXPro version 3.0: Expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2013, 41, D1206–D1213. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Jha, S.K.; Bagri, J.; Pandey, G.K. ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS ONE 2010, 5, e0125168. [Google Scholar] [CrossRef] [PubMed]
- Zang, G.; Zou, H.; Zhang, Y.; Xiang, Z.; Huang, J.; Luo, L.; Wang, C.; Lei, K.; Li, X.; Song, D.; et al. The De-Etiolated 1 homolog of Arabidopsis modulates the ABA signaling pathway and ABA biosynthesis in rice. Plant Physiol. 2016, 171, 1259–1276. [Google Scholar] [PubMed]
- Oono, Y.; Yazawa, T.; Kawahara, Y.; Kanamori, H.; Kobayashi, F.; Sasaki, H.; Mori, S.; Wu, J.; Handa, H.; Itoh, T.; et al. Genome-wide transcriptome analysis reveals that cadmium stress signaling controls the expression of genes in drought stress signal pathways in rice. PLoS ONE 2014, 9, e96946. [Google Scholar] [CrossRef] [PubMed]
- Willems, E.; Leyns, L.; Vandesompele, J. Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 2008, 379, 127–129. [Google Scholar] [CrossRef] [PubMed]









| Name | TIGR Locus ID | Chr. | Location | Open Reading Frame Length (bp) | Protein | Alias | ||
|---|---|---|---|---|---|---|---|---|
| Amino Acids (aa) | Molecular Weight (KD) | Isoelectric Point | ||||||
| OsPHD1 | LOC_Os01g08820 | 1 | 4431532–4433679 (R) | 1824 | 607 | 63.23 | 8.01 | OsPhd3 [18] |
| OsPHD2 | LOC_Os01g11952 | 1 | 6507356–6514891 (R) | 2976 | 991 | 110.89 | 8.18 | |
| OsPHD3 | LOC_Os01g11960 | 1 | 6518127–6521046 (R) | 1218 | 405 | 44.46 | 5.30 | |
| OsPHD4 | LOC_Os01g46700 | 1 | 26591955–26599634 (F) | 3048 | 1015 | 115.46 | 6.40 | |
| OsPHD5 | LOC_Os01g65600 | 1 | 38081608–38085548 (F) | 2151 | 716 | 79.23 | 7.44 | OsPhd4 [18] |
| OsPHD6 | LOC_Os01g66070 | 1 | 38344410–38344937 (F) | 528 | 175 | 19.75 | 4.84 | OsPhd5 [18] |
| OsPHD7 | LOC_Os01g66420 | 1 | 38566147–38570861(R) | 819 | 272 | 29.57 | 5.32 | OsPhd7 [18] |
| OsPHD8 | LOC_Os01g73460 | 1 | 42567843–42573082 (F) | 1158 | 385 | 43.48 | 8.92 | OsPhd2 [18] |
| OsPHD9 | LOC_Os01g73480 | 1 | 42579342–42583645 (F) | 2694 | 897 | 95.02 | 9.65 | |
| OsPHD10 | LOC_Os02g03030 | 2 | 1190846–1192461 (F) | 1086 | 361 | 39.59 | 9.16 | OsPhd11 [18] |
| OsPHD11 | LOC_Os02g09910 | 2 | 5131380–5132629 (F) | 780 | 259 | 26.93 | 5.78 | |
| OsPHD12 | LOC_Os02g09920 | 2 | 5145548–5151131 (F) | 2940 | 979 | 110.22 | 4.85 | |
| OsPHD13 | LOC_Os02g35600 | 2 | 21413884–21417221 (F) | 804 | 267 | 29.20 | 6.00 | OsPhd12 [18] |
| OsPHD14 | LOC_Os02g48810 | 2 | 29871368–29875355 (F) | 2064 | 687 | 71.93 | 5.25 | OsPhd14 [18] |
| OsPHD15 | LOC_Os02g52960 | 2 | 32384631–32396094 (F) | 4461 | 1486 | 164.12 | 7.48 | |
| OsPHD16 | LOC_Os03g04980 | 3 | 2409861–2413432 (F) | 762 | 253 | 27.61 | 5.65 | OsPhd18 [18] |
| OsPHD17 | LOC_Os03g19020 | 3 | 10651192–10655656 (F) | 2400 | 799 | 90.27 | 4.54 | |
| OsPHD18 | LOC_Os03g19190 | 3 | 10752594–10759931 (F) | 4389 | 1462 | 158.32 | 7.95 | |
| OsPHD19 | LOC_Os03g50780 | 3 | 28988380–28991862 (F) | 2094 | 697 | 76.64 | 5.86 | |
| OsPHD20 | LOC_Os03g53630 | 3 | 30747354–30755128 (F) | 4038 | 1345 | 146.86 | 6.70 | OsPhd17 [18] |
| OsPHD21 | LOC_Os03g53700 | 3 | 30791616–30797021 (F) | 762 | 253 | 28.77 | 4.66 | |
| OsPHD22 | LOC_Os03g58530 | 3 | 33328214–33333421 (F) | 657 | 218 | 25.02 | 8.34 | |
| OsPHD23 | LOC_Os03g60390 | 3 | 34342899–34346745 (R) | 744 | 247 | 27.75 | 5.64 | OsPhd16 [18] |
| OsPHD24 | LOC_Os04g34720 | 4 | 21001722–21010737 (F) | 1314 | 437 | 48.44 | 5.41 | OsPhd21 [18] |
| OsPHD25 | LOC_Os04g35430 | 4 | 21559229–21565988 (R) | 4308 | 1435 | 160.19 | 8.03 | OsPhd20 [18] |
| OsPHD26 | LOC_Os04g36730 | 4 | 22189014–22191831 (F) | 771 | 256 | 28.24 | 5.98 | |
| OsPHD27 | LOC_Os04g52020 | 4 | 30888156–30903920 (R) | 2439 | 812 | 90.94 | 6.60 | |
| OsPHD28 | LOC_Os04g59510 | 4 | 35382882–35390423 (R) | 1158 | 385 | 43.00 | 5.13 | |
| OsPHD29 | LOC_Os05g03430 | 5 | 1426929–1438465 (F) | 2631 | 876 | 97.00 | 4.81 | |
| OsPHD30 | LOC_Os05g07040 | 5 | 3697729–3702359 (R) | 777 | 258 | 28.84 | 5.39 | |
| OsPHD31 | LOC_Os05g34640 | 5 | 20542370–20547114 (F) | 777 | 258 | 29.07 | 5.28 | OsPhd22 [18] |
| OsPHD32 | LOC_Os06g01170 | 6 | 131089–138360 (F) | 3078 | 1025 | 111.11 | 6.70 | OsPhd24 [18] |
| OsPHD33 | LOC_Os06g08790 | 6 | 4393376–4401129 (F) | 2445 | 814 | 91.95 | 8.67 | |
| OsPHD34 | LOC_Os06g10690 | 6 | 5575867–5584373 (F) | 3420 | 1139 | 125.22 | 7.20 | |
| OsPHD35 | LOC_Os06g12400 | 6 | 6725306–6734446 (F) | 2379 | 792 | 87.21 | 4.95 | OsHAZ1 [27] |
| OsPHD36 | LOC_Os06g17280 | 6 | 10003546–10007033 (F) | 2292 | 763 | 83.42 | 7.58 | |
| OsPHD37 | LOC_Os06g20410 | 6 | 11725804–11734594 (R) | 1989 | 662 | 71.45 | 9.02 | OsPhd28 [18] |
| OsPHD38 | LOC_Os06g51450 | 6 | 31161699–31169513 (R) | 2400 | 799 | 87.56 | 9.87 | |
| OsPHD39 | LOC_Os06g51490 | 6 | 31184911–31187612 (R) | 864 | 287 | 32.52 | 7.39 | |
| OsPHD40 | LOC_Os07g07690 | 7 | 3859755–3868364 (F) | 4329 | 1442 | 156.86 | 8.46 | |
| OsPHD41 | LOC_Os07g08880 | 7 | 4606679–4614599 (F) | 657 | 218 | 24.89 | 7.94 | |
| OsPHD42 | LOC_Os07g12910 | 7 | 7414568–7418059 (R) | 735 | 244 | 27.37 | 6.39 | OsPhd34 [18] |
| OsPHD43 | LOC_Os07g31450 | 7 | 18625785–18638679 (R) | 6579 | 2192 | 243.66 | 6.37 | |
| OsPHD44 | LOC_Os07g41740 | 7 | 25013506–25017814 (F) | 837 | 278 | 30.38 | 5.56 | OsPhd32 [18] |
| OsPHD45 | LOC_Os07g46690 | 7 | 27902966–27912209 (R) | 5022 | 1673 | 186.85 | 6.80 | OsPhd30 [18] |
| OsPHD46 | LOC_Os07g49030 | 7 | 29353044–29356917 (R) | 2361 | 786 | 88.01 | 4.46 | OsPhd29 [18] |
| OsPHD47 | LOC_Os07g49290 | 7 | 29519451–29522145 (R) | 2235 | 744 | 83.32 | 8.32 | OsPhd31 [18] |
| OsPHD48 | LOC_Os08g01420 | 8 | 272854–276978 (R) | 1692 | 563 | 62.13 | 6.07 | OsPhd35 [18] OsEhd3 [28] |
| OsPHD49 | LOC_Os08g32620 | 8 | 20204623–20208055 (F) | 651 | 216 | 24.82 | 8.41 | |
| OsPHD50 | LOC_Os09g04890 | 9 | 2601650–2615934 (R) | 3069 | 1022 | 115.53 | 7.48 | |
| OsPHD51 | LOC_Os09g21770 | 9 | 13181330–13184741 (F) | 651 | 216 | 24.64 | 8.84 | |
| OsPHD52 | LOC_Os09g27620 | 9 | 16786041–16788255 (F) | 2040 | 679 | 73.76 | 7.60 | OsMS1 [29] |
| OsPHD53 | LOC_Os09g38440 | 9 | 22130437–22135245 (F) | 2133 | 710 | 79.43 | 9.16 | |
| OsPHD54 | LOC_Os11g05130 | 11 | 2247706–2256169 (R) | 5667 | 1888 | 206.55 | 5.70 | OsPhd37 [18] |
| OsPHD55 | LOC_Os11g12650 | 11 | 7125124–7128857 (R) | 2148 | 715 | 79.80 | 8.29 | |
| OsPHD56 | LOC_Os11g14010 | 11 | 7776646–7784194 (R) | 765 | 254 | 28.51 | 5.16 | OsPhd40 [18] |
| OsPHD57 | LOC_Os11g18770 | 11 | 10648362–10657255 (R) | 3171 | 1056 | 113.99 | 7.91 | |
| OsPHD58 | LOC_Os12g24540 | 12 | 14012791–14020313 (R) | 3807 | 1268 | 138.94 | 5.39 | |
| OsPHD59 | LOC_Os12g34330 | 12 | 20806080–20813363 (R) | 2067 | 688 | 76.07 | 9.22 | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, M.; Jia, B.; Yang, J.; Cui, N.; Zhu, Y.; Sun, X. Genome-Wide Identification of the PHD-Finger Family Genes and Their Responses to Environmental Stresses in Oryza sativa L. Int. J. Mol. Sci. 2017, 18, 2005. https://doi.org/10.3390/ijms18092005
Sun M, Jia B, Yang J, Cui N, Zhu Y, Sun X. Genome-Wide Identification of the PHD-Finger Family Genes and Their Responses to Environmental Stresses in Oryza sativa L. International Journal of Molecular Sciences. 2017; 18(9):2005. https://doi.org/10.3390/ijms18092005
Chicago/Turabian StyleSun, Mingzhe, Bowei Jia, Junkai Yang, Na Cui, Yanming Zhu, and Xiaoli Sun. 2017. "Genome-Wide Identification of the PHD-Finger Family Genes and Their Responses to Environmental Stresses in Oryza sativa L." International Journal of Molecular Sciences 18, no. 9: 2005. https://doi.org/10.3390/ijms18092005
APA StyleSun, M., Jia, B., Yang, J., Cui, N., Zhu, Y., & Sun, X. (2017). Genome-Wide Identification of the PHD-Finger Family Genes and Their Responses to Environmental Stresses in Oryza sativa L. International Journal of Molecular Sciences, 18(9), 2005. https://doi.org/10.3390/ijms18092005