How Sweet Are Our Gut Beneficial Bacteria? A Focus on Protein Glycosylation in Lactobacillus
Abstract
:1. Introduction
2. Overview of Protein Glycosylation in Prokaryotes
2.1. N-Glycosylation: The Campylobacter jejuni Paradigm
2.2. Alternative N-Glycosylation in β- and γ-Proteobacteria
2.3. N-Glycosylation in Mycoplasmas
2.4. O-Glycosylation in Bacteria
2.5. En Bloc O-Glycosylation
2.6. O-Glycosylation by Sequential Action of Glycosyltransferases
2.7. The Accessory Secretion System SecA2 Glycosylation Pathway
3. Protein Glycosylation in Gut Commensal Bacteria
4. Protein Glycosylation in Lactobacillus
4.1. Lactobacillus Glycoproteins
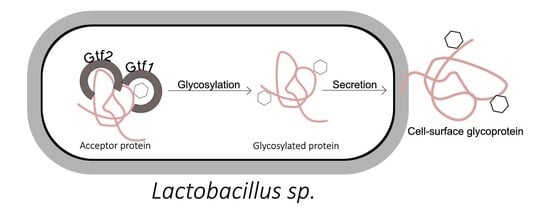
4.2. Protein Glycosylation Pathways in Lactobacillus
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bastos, P.A.D.; da Costa, J.P.; Vitorino, R. A glimpse into the modulation of post-translational modifications of human-colonizing bacteria. J. Proteom. 2017, 152, 254–275. [Google Scholar] [CrossRef] [PubMed]
- Dell, A.; Galadari, A.; Sastre, F.; Hitchen, P. Similarities and differences in the glycosylation mechanisms in prokaryotes and eukaryotes. Int. J. Microbiol. 2010, 2010, 148178. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.; Jarrell, H.; Millar, L.; Tessier, L.; Fiori, L.M.; Lau, P.C.; Allan, B.; Szymanski, C.M. Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer. J. Bacteriol. 2006, 188, 2427–2434. [Google Scholar] [CrossRef] [PubMed]
- Linton, D.; Dorrell, N.; Hitchen, P.G.; Amber, S.; Karlyshev, A.V.; Morris, H.R.; Dell, A.; Valvano, M.A.; Aebi, M.; Wren, B.W. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol. Microbiol. 2005, 55, 1695–1703. [Google Scholar] [CrossRef] [PubMed]
- Young, N.M.; Brisson, J.-R.; Kelly, J.; Watson, D.C.; Tessier, L.; Lanthier, P.H.; Jarrell, H.C.; Cadotte, N.; St Michael, F.; Aberg, E.; et al. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 2002, 277, 42530–42539. [Google Scholar] [CrossRef] [PubMed]
- Nothaft, H.; Scott, N.E.; Vinogradov, E.; Liu, X.; Hu, R.; Beadle, B.; Fodor, C.; Miller, W.G.; Li, J.; Cordwell, S.J.; et al. Diversity in the protein N-glycosylation pathways within the Campylobacter genus. Mol. Cell. Proteom. 2012, 11, 1203–1219. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.F.; Wacker, M.; Hernandez, M.; Hitchen, P.G.; Marolda, C.L.; Kowarik, M.; Morris, H.R.; Dell, A.; Valvano, M.A.; Aebi, M. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc. Natl. Acad. Sci. USA 2005, 102, 3016–3021. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, F.; Huang, W.; Li, C.; Schulz, B.L.; Lizak, C.; Palumbo, A.; Numao, S.; Neri, D.; Aebi, M.; Wang, L.-X. A combined method for producing homogeneous glycoproteins with eukaryotic N-glycosylation. Nat. Chem. Biol. 2010, 6, 264–266. [Google Scholar] [CrossRef] [PubMed]
- Valderrama-Rincon, J.D.; Fisher, A.C.; Merritt, J.H.; Fan, Y.-Y.; Reading, C.A.; Chhiba, K.; Heiss, C.; Azadi, P.; Aebi, M.; DeLisa, M.P. An engineered eukaryotic protein glycosylation pathway in Escherichia coli. Nat. Chem. Biol. 2012, 8, 434–436. [Google Scholar] [CrossRef] [PubMed]
- Grass, S.; Lichti, C.F.; Townsend, R.R.; Gross, J.; St Geme, J.W. The Haemophilus influenzae HMW1C protein is a glycosyltransferase that transfers hexose residues to asparagine sites in the HMW1 adhesin. PLoS Pathog. 2010, 6, e1000919. [Google Scholar] [CrossRef] [PubMed]
- Gross, J.; Grass, S.; Davis, A.E.; Gilmore-Erdmann, P.; Townsend, R.R.; St Geme, J.W. The Haemophilus influenzae HMW1 adhesin is a glycoprotein with an unusual N-linked carbohydrate modification. J. Biol. Chem. 2008, 283, 26010–26015. [Google Scholar] [CrossRef] [PubMed]
- McCann, J.R.; St Geme, J.W. The HMW1C-like glycosyltransferase—An enzyme family with a sweet tooth for simple sugars. PLoS Pathog. 2014, 10, e1003977. [Google Scholar] [CrossRef] [PubMed]
- Naegeli, A.; Neupert, C.; Fan, Y.-Y.; Lin, C.-W.; Poljak, K.; Papini, A.M.; Schwarz, F.; Aebi, M. Molecular analysis of an alternative N-glycosylation machinery by functional transfer from Actinobacillus pleuropneumoniae to Escherichia coli. J. Biol. Chem. 2014, 289, 2170–2179. [Google Scholar] [CrossRef] [PubMed]
- Naegeli, A.; Michaud, G.; Schubert, M.; Lin, C.-W.; Lizak, C.; Darbre, T.; Reymond, J.-L.; Aebi, M. Substrate specificity of cytoplasmic N-glycosyltransferase. J. Biol. Chem. 2014, 289, 24521–24532. [Google Scholar] [CrossRef] [PubMed]
- Rempe, K.A.; Spruce, L.A.; Porsch, E.A.; Seeholzer, S.H.; Nørskov-Lauritsen, N.; St Geme, J.W. Unconventional N-linked glycosylation promotes trimeric autotransporter function in kingella kingae and Aggregatibacter aphrophilus. MBio 2015, 6, e01206-15. [Google Scholar] [CrossRef] [PubMed]
- Cuccui, J.; Terra, V.S.; Bossé, J.T.; Naegeli, A.; Abouelhadid, S.; Li, Y.; Lin, C.W.; Vohra, P.; Tucker, A.W.; Rycroft, A.N.; et al. The N-linking glycosylation system from Actinobacillus pleuropneumoniae is required for adhesion and has potential use in glycoengineering. Open Biol. 2017, 7, 160212. [Google Scholar] [CrossRef] [PubMed]
- Daubenspeck, J.M.; Jordan, D.S.; Simmons, W.; Renfrow, M.B.; Dybvig, K. General N-and O-linked glycosylation of lipoproteins in mycoplasmas and role of exogenous oligosaccharide. PLoS ONE 2015, 10, e0143362. [Google Scholar] [CrossRef] [PubMed]
- Tan, F.Y.Y.; Tang, C.M.; Exley, R.M. Sugar coating: Bacterial protein glycosylation and host-microbe interactions. Trends Biochem. Sci. 2015, 40, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Schäffer, C.; Messner, P. Emerging facets of prokaryotic glycosylation. FEMS Microbiol. Rev. 2017, 41, 49–91. [Google Scholar] [CrossRef] [PubMed]
- Aas, F.E.; Vik, A.; Vedde, J.; Koomey, M.; Egge-Jacobsen, W. Neisseria gonorrhoeae O-linked pilin glycosylation: Functional analyses define both the biosynthetic pathway and glycan structure. Mol. Microbiol. 2007, 65, 607–624. [Google Scholar] [CrossRef] [PubMed]
- Hartley, M.D.; Morrison, M.J.; Aas, F.E.; Børud, B.; Koomey, M.; Imperiali, B. Biochemical characterization of the O-linked glycosylation pathway in Neisseria gonorrhoeae responsible for biosynthesis of protein glycans containing N,N′-diacetylbacillosamine. Biochemistry 2011, 50, 4936–4948. [Google Scholar] [CrossRef] [PubMed]
- Vik, A.; Aas, F.E.; Anonsen, J.H.; Bilsborough, S.; Schneider, A.; Egge-Jacobsen, W.; Koomey, M. Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 2009, 106, 4447–4452. [Google Scholar] [CrossRef] [PubMed]
- Cuccui, J.; Wren, B.W. Bacteria like sharing their sweets. Mol. Microbiol. 2013, 89, 811–815. [Google Scholar] [CrossRef] [PubMed]
- DiGiandomenico, A.; Matewish, M.J.; Bisaillon, A.; Stehle, J.R.; Lam, J.S.; Castric, P. Glycosylation of Pseudomonas aeruginosa 1244 pilin: Glycan substrate specificity. Mol. Microbiol. 2002, 46, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Horzempa, J.; Dean, C.R.; Goldberg, J.B.; Castric, P. Pseudomonas aeruginosa 1244 pilin glycosylation: Glycan substrate recognition. J. Bacteriol. 2006, 188, 4244–4252. [Google Scholar] [CrossRef] [PubMed]
- Balonova, L.; Mann, B.F.; Cerveny, L.; Alley, W.R.; Chovancova, E.; Forslund, A.-L.; Salomonsson, E.N.; Forsberg, A.; Damborsky, J.; Novotny, M.V.; et al. Characterization of protein glycosylation in Francisella tularensis subsp. holarctica: Identification of a novel glycosylated lipoprotein required for virulence. Mol. Cell. Proteom. 2012, 11, M111–015016. [Google Scholar] [CrossRef] [PubMed]
- Samuel, G.; Reeves, P. Biosynthesis of O-antigens: Genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 2003, 338, 2503–2519. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.M.; Nasr, M.A.; Kinsella, R.L.; Scott, N.E.; Foster, L.J.; Weber, B.S.; Fiester, S.E.; Actis, L.A.; Tracy, E.N.; Munson, R.S.; et al. Acinetobacter strains carry two functional oligosaccharyltransferases, one devoted exclusively to type IV pilin, and the other one dedicated to O-glycosylation of multiple proteins. Mol. Microbiol. 2015, 96, 1023–1041. [Google Scholar] [CrossRef] [PubMed]
- Anonsen, J.H.; Vik, Å.; Børud, B.; Viburiene, R.; Aas, F.E.; Kidd, S.W.A.; Aspholm, M.; Koomey, M. Characterization of a unique tetrasaccharide and distinct glycoproteome in the O-linked protein glycosylation System of Neisseria elongata subsp. glycolytica. J. Bacteriol. 2015, 198, 256–267. [Google Scholar] [CrossRef] [PubMed]
- John, C.M.; Liu, M.; Phillips, N.J.; Yang, Z.; Funk, C.R.; Zimmerman, L.I.; Griffiss, J.M.; Stein, D.C.; Jarvis, G.A. Lack of lipid A pyrophosphorylation and functional lptA reduces inflammation by Neisseria commensals. Infect. Immun. 2012, 80, 4014–4026. [Google Scholar] [CrossRef] [PubMed]
- Lithgow, K.V.; Scott, N.E.; Iwashkiw, J.A.; Thomson, E.L.S.; Foster, L.J.; Feldman, M.F.; Dennis, J.J. A general protein O-glycosylation system within the Burkholderia cepacia complex is involved in motility and virulence. Mol. Microbiol. 2014, 92, 116–137. [Google Scholar] [CrossRef] [PubMed]
- Qutyan, M.; Henkel, M.; Horzempa, J.; Quinn, M.; Castric, P. Glycosylation of pilin and nonpilin protein constructs by Pseudomonas aeruginosa 1244. J. Bacteriol. 2010, 192, 5972–5981. [Google Scholar] [CrossRef] [PubMed]
- Zampronio, C.G.; Blackwell, G.; Penn, C.W.; Cooper, H.J. Novel glycosylation sites localized in Campylobacter jejuni flagellin FlaA by liquid chromatography electron capture dissociation tandem mass spectrometry. J. Proteome Res. 2011, 10, 1238–1245. [Google Scholar] [CrossRef] [PubMed]
- Zebian, N.; Merkx-Jacques, A.; Pittock, P.P.; Houle, S.; Dozois, C.M.; Lajoie, G.A.; Creuzenet, C. Comprehensive analysis of flagellin glycosylation in Campylobacter jejuni NCTC 11168 reveals incorporation of legionaminic acid and its importance for host colonization. Glycobiology 2016, 26, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Twine, S.M.; Paul, C.J.; Vinogradov, E.; McNally, D.J.; Brisson, J.-R.; Mullen, J.A.; McMullin, D.R.; Jarrell, H.C.; Austin, J.W.; Kelly, J.F.; et al. Flagellar glycosylation in Clostridium botulinum. FEBS J. 2008, 275, 4428–4444. [Google Scholar] [CrossRef] [PubMed]
- Faulds-Pain, A.; Twine, S.M.; Vinogradov, E.; Strong, P.C.R.; Dell, A.; Buckley, A.M.; Douce, G.R.; Valiente, E.; Logan, S.M.; Wren, B.W. The post-translational modification of the Clostridium difficile flagellin affects motility, cell surface properties and virulence. Mol. Microbiol. 2014, 94, 272–289. [Google Scholar] [CrossRef] [PubMed]
- Twine, S.M.; Reid, C.W.; Aubry, A.; McMullin, D.R.; Fulton, K.M.; Austin, J.; Logan, S.M. Motility and flagellar glycosylation in Clostridium difficile. J. Bacteriol. 2009, 191, 7050–7062. [Google Scholar] [CrossRef] [PubMed]
- Bouché, L.; Panico, M.; Hitchen, P.; Binet, D.; Sastre, F.; Faulds-Pain, A.; Valiente, E.; Vinogradov, E.; Aubry, A.; Fulton, K.; et al. The Type B flagellin of hypervirulent Clostridium difficile is modified with novel sulfonated peptidylamido-glycans. J. Biol. Chem. 2016, 291, 25439–25449. [Google Scholar] [CrossRef] [PubMed]
- Valiente, E.; Bouché, L.; Hitchen, P.; Faulds-Pain, A.; Songane, M.; Dawson, L.F.; Donahue, E.; Stabler, R.A.; Panico, M.; Morris, H.R.; et al. Role of glycosyltransferases modifying type B flagellin of emerging hypervirulent clostridium difficile lineages and their impact on motility and biofilm formation. J. Biol. Chem. 2016, 291, 25450–25461. [Google Scholar] [CrossRef] [PubMed]
- Maes, E.; Krzewinski, F.; Garenaux, E.; Lequette, Y.; Coddeville, B.; Trivelli, X.; Ronse, A.; Faille, C.; Guerardel, Y. Glycosylation of BclA glycoprotein from Bacillus cereus and Bacillus anthracis exosporium is domain-specific. J. Biol. Chem. 2016, 291, 9666–9677. [Google Scholar] [CrossRef] [PubMed]
- Schirm, M.; Kalmokoff, M.; Aubry, A.; Thibault, P.; Sandoz, M.; Logan, S.M. Flagellin from Listeria monocytogenes is glycosylated with beta-O-linked N-acetylglucosamine. J. Bacteriol. 2004, 186, 6721–6727. [Google Scholar] [CrossRef] [PubMed]
- Hart, G.W.; Slawson, C.; Ramirez-Correa, G.; Lagerlof, O. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2011, 80, 825–858. [Google Scholar] [CrossRef] [PubMed]
- Prabudiansyah, I.; Driessen, A.J.M. The Canonical and accessory Sec system of gram-positive bacteria. Curr. Top. Microbiol. Immunol. 2017, 404, 45–67. [Google Scholar] [CrossRef] [PubMed]
- Lizcano, A.; Sanchez, C.J.; Orihuela, C.J. A role for glycosylated serine-rich repeat proteins in gram-positive bacterial pathogenesis. Mol. Oral Microbiol. 2012, 27, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Bu, S.; Li, Y.; Zhou, M.; Azadin, P.; Zeng, M.; Fives-Taylor, P.; Wu, H. Interaction between two putative glycosyltransferases is required for glycosylation of a serine-rich streptococcal adhesin. J. Bacteriol. 2008, 190, 1256–1266. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Sun, B.; Wu, H.; Peng, Z.; Fives-Taylor, P.M. Differential roles of individual domains in selection of secretion route of a Streptococcus parasanguinis serine-rich adhesin, Fap1. J. Bacteriol. 2007, 189, 7610–7617. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Wu, H.; Ruiz, T.; Chen, Q.; Zhou, M.; Sun, B.; Fives-Taylor, P. Role of gap3 in Fap1 glycosylation, stability, in vitro adhesion, and fimbrial and biofilm formation of Streptococcus parasanguinis. Oral Microbiol. Immunol. 2008, 23, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhou, M.; Yang, T.; Haslam, S.M.; Dell, A.; Wu, H. A new helical binding domain mediates a unique glycosyltransferase activity of a bifunctional protein. J. Biol Chem. 2016, 291, 22106–22117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, F.; Yang, T.; Ding, L.; Zhou, M.; Li, J.; Haslam, S.M.; Dell, A.; Erlandsen, H.; Wu, H. The highly conserved domain of unknown function 1792 has a distinct glycosyltransferase fold. Nat. Commun. 2014, 5, 4339. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zhu, F.; Dong, S.; Pritchard, D.G.; Wu, H. A novel glucosyltransferase is required for glycosylation of a serine-rich adhesin and biofilm formation by Streptococcus parasanguinis. J. Biol. Chem. 2010, 285, 12140–12148. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Erlandsen, H.; Ding, L.; Li, J.; Huang, Y.; Zhou, M.; Liang, X.; Ma, J.; Wu, H. Structural and functional analysis of a new subfamily of glycosyltransferases required for glycosylation of serine-rich streptococcal adhesins. J. Biol. Chem. 2011, 286, 27048–27057. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Zhang, H.; Yang, T.; Haslam, S.M.; Dell, A.; Wu, H. Engineering and dissecting the glycosylation pathway of a streptococcal serine-rich repeat adhesin. J. Biol. Chem. 2016, 291, 27354–27363. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Seepersaud, R.; Bensing, B.A.; Sullam, P.M.; Rapoport, T.A. Mechanism of a cytosolic O-glycosyltransferase essential for the synthesis of a bacterial adhesion protein. Proc. Natl. Acad. Sci. USA 2016, 113, E1190–E1199. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.-W.; Jiang, Y.-L.; Zhu, F.; Yang, Y.-H.; Shao, Q.-Y.; Yang, H.-B.; Ren, Y.-M.; Wu, H.; Chen, Y.; Zhou, C.-Z. Structure of a novel O-linked N-acetyl-d-glucosamine (O-GlcNAc) transferase, GtfA, reveals insights into the glycosylation of pneumococcal serine-rich repeat adhesins. J. Biol. Chem. 2014, 289, 20898–20907. [Google Scholar] [CrossRef] [PubMed]
- Lizcano, A.; Babu, R.A.S.; Shenoy, A.T.; Saville, A.M.; Kumar, N.; D’Mello, A.; Hinojosa, C.A.; Gilley, R.P.; Segovia, J.; Mitchell, T.J.; et al. Transcriptional organization of pneumococcal psrP-secY2A2 and impact of GtfA and GtfB deletion on PsrP-associated virulence properties. Microbes Infect. 2017, 19, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Mistou, M.-Y.; Dramsi, S.; Brega, S.; Poyart, C.; Trieu-Cuot, P. Molecular dissection of the secA2 locus of group B Streptococcus reveals that glycosylation of the Srr1 LPXTG protein is required for full virulence. J. Bacteriol. 2009, 191, 4195–4206. [Google Scholar] [CrossRef] [PubMed]
- Valguarnera, E.; Kinsella, R.L.; Feldman, M.F. Sugar and spice make bacteria not nice: Protein glycosylation and its influence in pathogenesis. J. Mol. Biol. 2016, 428, 3206–3220. [Google Scholar] [CrossRef] [PubMed]
- Coyne, M.J.; Reinap, B.; Lee, M.M.; Comstock, L.E. Human symbionts use a host-like pathway for surface fucosylation. Science 2005, 307, 1778–1781. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, C.M.; Coyne, M.J.; Villa, O.F.; Chatzidaki-Livanis, M.; Comstock, L.E. A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell 2009, 137, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, C.M.; Coyne, M.J.; Comstock, L.E. Theoretical and experimental characterization of the scope of protein O-glycosylation in Bacteroides fragilis. J. Biol. Chem. 2011, 286, 3219–3226. [Google Scholar] [CrossRef] [PubMed]
- Couvigny, B.; Lapaque, N.; Rigottier-Gois, L.; Guillot, A.; Chat, S.; Meylheuc, T.; Kulakauskas, S.; Rohde, M.; Mistou, M.-Y.; Renault, P.; et al. Three glycosylated serine-rich repeat proteins play a pivotal role in adhesion and colonization of the pioneer commensal bacterium, Streptococcus salivarius. Environ. Microbiol. 2017, 19, 3579–3594. [Google Scholar] [CrossRef] [PubMed]
- Fredriksen, L.; Mathiesen, G.; Moen, A.; Bron, P.A.; Kleerebezem, M.; Eijsink, V.G.H.; Egge-Jacobsen, W. The major autolysin Acm2 from Lactobacillus plantarum undergoes cytoplasmic O-glycosylation. J. Bacteriol. 2012, 194, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Rolain, T.; Bernard, E.; Beaussart, A.; Degand, H.; Courtin, P.; Egge-Jacobsen, W.; Bron, P.A.; Morsomme, P.; Kleerebezem, M.; Chapot-Chartier, M.-P.; et al. O-glycosylation as a novel control mechanism of peptidoglycan hydrolase activity. J. Biol. Chem. 2013, 288, 22233–22247. [Google Scholar] [CrossRef] [PubMed]
- Lebeer, S.; Claes, I.J.J.; Balog, C.I.A.; Schoofs, G.; Verhoeven, T.L.A.; Nys, K.; von Ossowski, I.; de Vos, W.M.; Tytgat, H.L.P.; Agostinis, P.; et al. The major secreted protein Msp1/p75 is O-glycosylated in Lactobacillus rhamnosus GG. Microb. Cell Fact. 2012, 11, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anzengruber, J.; Pabst, M.; Neumann, L.; Sekot, G.; Heinl, S.; Grabherr, R.; Altmann, F.; Messner, P.; Schäffer, C. Protein O-glucosylation in Lactobacillus buchneri. Glycoconj. J. 2014, 31, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Juge, N. Microbial adhesins to gastrointestinal mucus. Trends Microbiol. 2012, 20, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Tytgat, H.L.P.; van Teijlingen, N.H.; Sullan, R.M.A.; Douillard, F.P.; Rasinkangas, P.; Messing, M.; Reunanen, J.; Satokari, R.; Vanderleyden, J.; Dufrêne, Y.F.; et al. Probiotic gut microbiota isolate interacts with dendritic cells via glycosylated heterotrimeric Pili. PLoS ONE 2016, 11, e0151824. [Google Scholar] [CrossRef] [PubMed]
- Kajikawa, A.; Midorikawa, E.; Masuda, K.; Kondo, K.; Irisawa, T.; Igimi, S.; Okada, S. Characterization of flagellins isolated from a highly motile strain of Lactobacillus agilis. BMC Microbiol. 2016, 16, 49. [Google Scholar] [CrossRef] [PubMed]
- Mobili, P.; de los Ángeles Serradell, M.; Trejo, S.A.; Avilés Puigvert, F.X.; Abraham, A.G.; De Antoni, G.L. Heterogeneity of S-layer proteins from aggregating and non-aggregating Lactobacillus kefir strains. Antonie Leeuwenhoek 2009, 95, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Fredriksen, L.; Moen, A.; Adzhubei, A.A.; Mathiesen, G.; Eijsink, V.G.H.; Egge-Jacobsen, W. Lactobacillus plantarum WCFS1 O-linked protein glycosylation: An extended spectrum of target proteins and modification sites detected by mass spectrometry. Glycobiology 2013, 23, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- Nikolic, M.; López, P.; Strahinic, I.; Suárez, A.; Kojic, M.; Fernández-García, M.; Topisirovic, L.; Golic, N.; Ruas-Madiedo, P. Characterisation of the exopolysaccharide (EPS)-producing Lactobacillus paraplantarum BGCG11 and its non-EPS producing derivative strains as potential probiotics. Int. J. Food Microbiol. 2012, 158, 155–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sengupta, R.; Altermann, E.; Anderson, R.C.; McNabb, W.C.; Moughan, P.J.; Roy, N.C. The role of cell surface architecture of lactobacilli in host-microbe interactions in the gastrointestinal tract. Mediat. Inflamm. 2013, 2013, 237921. [Google Scholar] [CrossRef] [PubMed]
- Kankainen, M.; Paulin, L.; Tynkkynen, S.; von Ossowski, I.; Reunanen, J.; Partanen, P.; Satokari, R.; Vesterlund, S.; Hendrickx, A.P.A.; Lebeer, S.; et al. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc. Natl. Acad. Sci. USA 2009, 106, 17193–17198. [Google Scholar] [CrossRef] [PubMed]
- Lebeer, S.; Claes, I.; Tytgat, H.L.P.; Verhoeven, T.L.A.; Marien, E.; von Ossowski, I.; Reunanen, J.; Palva, A.; Vos, W.M.; de Keersmaecker, S.C.J.D.; et al. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 2012, 78, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Erdem, A.L.; Avelino, F.; Xicohtencatl-Cortes, J.; Girón, J.A. Host protein binding and adhesive properties of H6 and H7 flagella of attaching and effacing Escherichia coli. J. Bacteriol. 2007, 189, 7426–7435. [Google Scholar] [CrossRef] [PubMed]
- Guerry, P. Campylobacter flagella: Not just for motility. Trends Microbiol. 2007, 15, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Neville, B.A.; Forde, B.M.; Claesson, M.J.; Darby, T.; Coghlan, A.; Nally, K.; Ross, R.P.; O’Toole, P.W. Characterization of pro-inflammatory flagellin proteins produced by Lactobacillus ruminis and related motile Lactobacilli. PLoS ONE 2012, 7, e40592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hynönen, U.; Palva, A. Lactobacillus surface layer proteins: Structure, function and applications. Appl. Microbiol. Biotechnol. 2013, 97, 5225–5243. [Google Scholar] [CrossRef] [PubMed]
- Sleytr, U.B.; Schuster, B.; Egelseer, E.-M.; Pum, D. S-layers: Principles and applications. FEMS Microbiol. Rev. 2014, 38, 823–864. [Google Scholar] [CrossRef] [PubMed]
- Avall-Jääskeläinen, S.; Palva, A. Lactobacillus surface layers and their applications. FEMS Microbiol. Rev. 2005, 29, 511–529. [Google Scholar] [CrossRef] [PubMed]
- Mozes, N.; Lortal, S. X-ray photoelectron spectroscopy and biochemical analysis of the surface of Lactobacillus helveticus ATCC 12046. Microbiology 1995, 141, 11–19. [Google Scholar] [CrossRef]
- Möschl, A.; Schäffer, C.; Sleytr, U.B.; Messner, P.; Christian, R.; Schulz, G. Characterization of the S-Layer Glycoproteins of Two Lactobacilli. In Advances in Bacterial Paracrystalline Surface Layers; Beveridge, T.J., Koval, S.F., Eds.; Springer: Boston, MA, USA, 1993; pp. 281–284. [Google Scholar]
- Konstantinov, S.R.; Smidt, H.; de Vos, W.M.; Bruijns, S.C.M.; Singh, S.K.; Valence, F.; Molle, D.; Lortal, S.; Altermann, E.; Klaenhammer, T.R.; et al. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc. Natl. Acad. Sci. USA 2008, 105, 19474–19479. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, D.A.; Tailford, L.E.; Hemmings, A.M.; Juge, N. Crystal structure of a mucus-binding protein repeat reveals an unexpected functional immunoglobulin binding activity. J. Biol. Chem. 2009, 284, 32444–32453. [Google Scholar] [CrossRef] [PubMed]
- Roos, S.; Jonsson, H. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 2002, 148, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Etzold, S.; Kober, O.I.; Mackenzie, D.A.; Tailford, L.E.; Gunning, A.P.; Walshaw, J.; Hemmings, A.M.; Juge, N. Structural basis for adaptation of lactobacilli to gastrointestinal mucus. Environ. Microbiol. 2014, 16, 888–903. [Google Scholar] [CrossRef] [PubMed]
- Gunning, A.P.; Kavanaugh, D.; Thursby, E.; Etzold, S.; MacKenzie, D.A.; Juge, N. Use of Atomic Force Microscopy to Study the Multi-Modular Interaction of Bacterial Adhesins to Mucins. Int. J. Mol. Sci. 2016, 17, 1854. [Google Scholar] [CrossRef] [PubMed]
- Etzold, S.; Juge, N. Structural insights into bacterial recognition of intestinal mucins. Curr. Opin. Struct. Biol. 2014, 28, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Bene, K.P.; Kavanaugh, D.W.; Leclaire, C.; Gunning, A.P.; MacKenzie, D.A.; Wittmann, A.; Young, I.D.; Kawasaki, N.; Rajnavolgyi, E.; Juge, N. Lactobacillus reuteri surface mucus adhesins upregulate inflammatory responses through interactions with innate C-type lectin receptors. Front. Microbiol. 2017, 8, 321. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.-C.; van Swam, I.I.; Tomita, S.; Morsomme, P.; Rolain, T.; Hols, P.; Kleerebezem, M.; Bron, P.A. GtfA and GtfB are both required for protein O-glycosylation in Lactobacillus plantarum. J. Bacteriol. 2014, 196, 1671–1682. [Google Scholar] [CrossRef] [PubMed]
- Wegmann, U.; MacKenzie, D.A.; Zheng, J.; Goesmann, A.; Roos, S.; Swarbreck, D.; Walter, J.; Crossman, L.C.; Juge, N. The pan-genome of Lactobacillus reuteri strains originating from the pig gastrointestinal tract. BMC Genom. 2015, 16, 1023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigel, N.W.; Braunstein, M. A new twist on an old pathway—Accessory secretion systems. Mol. Microbiol. 2008, 69, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Frese, S.A.; Benson, A.K.; Tannock, G.W.; Loach, D.M.; Kim, J.; Zhang, M.; Oh, P.L.; Heng, N.C.K.; Patil, P.B.; Juge, N.; et al. The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet. 2011, 7, e1001314. [Google Scholar] [CrossRef] [PubMed]
- Frese, S.A.; Mackenzie, D.A.; Peterson, D.A.; Schmaltz, R.; Fangman, T.; Zhou, Y.; Zhang, C.; Benson, A.K.; Cody, L.A.; Mulholland, F.; et al. Molecular characterization of host-specific biofilm formation in a vertebrate gut symbiont. PLoS Genet. 2013, 9, e1004057. [Google Scholar] [CrossRef] [PubMed]





| Protein | Organism | Glycan | Method * | Reference |
|---|---|---|---|---|
| Msp1 | L. rhamnosus GG | Man-containing | Pro-Q Emerald stain, Lectin affinity (WB, AFM) MS | [64] |
| SpaCBA | L. rhamnosus GG | Man and Fuc-containing | Lectin affinity (AFM, WB, ELLA) | [67] |
| FliC1/FliC2 | L. agilis | uncharacterized | PAS-stain | [68] |
| SlpB/N | L. buchneri CD034, L. buchneri NRRL B-30929 | Glc1–Glc7 | MS | [65] |
| LbGH25B/N Putative glycosyl-hydrolase | L. buchneri CD034, L. buchneri NRRL B-30929 | Glc8 | MS | [65] |
| Slp | L. kefir | uncharacterized | PAS stain | [69] |
| Acm2 | L. plantarum WCFS1 | GlcNAc | MS, lectin affinity (WB) | [62] |
| DnaK | L. plantarum WCFS1 | GlcNAc1, GlcNAc1Hex1 | MS | [70] |
| Lp_2162 (muropeptidase) | L. plantarum WCFS1 | GlcNAc1 | MS | [70] |
| Lp_2260 | L. plantarum WCFS1 | GlcNAc1 | MS | [70] |
| Lp_1643 (mucus binding protein) | L. plantarum WCFS1 | GlcNAc1 | MS | [70] |
| PdhC | L. plantarum WCFS1 | GlcNAc1 | MS | [70] |
| FtsY | L. plantarum WCFS1 | GlcNAc1 | MS | [70] |
| Lp_2793 | L. plantarum WCFS1 | GlcNAc1 | MS | [70] |
| FtsK1 | L. plantarum WCFS1 | GlcNAc1 | MS | [70] |
| Lp_3421 (muropeptidase) | L. plantarum WCFS1 | GlcNAc1, GlcNAc1Hex1 | MS | [70] |
| FtsZ | L. plantarum WCFS1 | GlcNAc1 | MS | [70] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Latousakis, D.; Juge, N. How Sweet Are Our Gut Beneficial Bacteria? A Focus on Protein Glycosylation in Lactobacillus. Int. J. Mol. Sci. 2018, 19, 136. https://doi.org/10.3390/ijms19010136
Latousakis D, Juge N. How Sweet Are Our Gut Beneficial Bacteria? A Focus on Protein Glycosylation in Lactobacillus. International Journal of Molecular Sciences. 2018; 19(1):136. https://doi.org/10.3390/ijms19010136
Chicago/Turabian StyleLatousakis, Dimitrios, and Nathalie Juge. 2018. "How Sweet Are Our Gut Beneficial Bacteria? A Focus on Protein Glycosylation in Lactobacillus" International Journal of Molecular Sciences 19, no. 1: 136. https://doi.org/10.3390/ijms19010136
APA StyleLatousakis, D., & Juge, N. (2018). How Sweet Are Our Gut Beneficial Bacteria? A Focus on Protein Glycosylation in Lactobacillus. International Journal of Molecular Sciences, 19(1), 136. https://doi.org/10.3390/ijms19010136